Publications
Publications in peer reviewed journals
Nitrogen kinetic isotope effects of nitrification by the complete ammonia oxidizer Nitrospira inopinata
2021 - mSphere, 6: e00634-21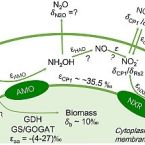
Abstract:
Analysis of nitrogen isotope fractionation effects is useful for tracing biogeochemical nitrogen cycle processes. Nitrification can cause large nitrogen isotope effects through the enzymatic oxidation of ammonia (NH3) via nitrite (NO2-) to nitrate (NO3-) (15εNH4+→NO2- and 15ɛNO2-→NO3-). The isotope effects of ammonia-oxidizing bacteria (AOB) and archaea (AOA), and nitrite-oxidizing bacteria (NOB), have been analyzed previously. Here we studied the nitrogen isotope effects of the complete ammonia oxidizer (comammox) Nitrospira inopinata that oxidizes NH3 to NO3-. At high ammonium (NH4+) availability (1 mM) and pH between 6.5 and 8.5, its 15εNH4+→NO2- ranged from −33.1 to −27.1‰ based on substrate consumption (residual substrate isotopic composition) and −35.5 to −31.2‰ based on product formation (cumulative product isotopic composition), while the 15ɛNO2-→NO3- ranged from 6.5 to 11.1‰ based on substrate consumption. These values resemble isotope effects of AOB and AOA, and of NOB in the genus Nitrospira, suggesting the absence of fundamental mechanistic differences between key enzymes for ammonia and nitrite oxidation in comammox and canonical nitrifiers. However, ambient pH and initial NH4+ concentrations influenced the isotope effects in N. inopinata. The 15εNH4+→NO2- based on product formation was smaller at pH 6.5 (−31.2‰) compared to pH 7.5 (−35.5‰) and pH 8.5 (−34.9‰), while 15ɛNO2-→NO3- was smaller at pH 8.5 (6.5‰) compared to pH 7.5 (8.8‰) and pH 6.5 (11.1‰). Isotopic fractionation via 15εNH4+→NO2- and 15ɛNO2-→NO3- was smaller at 0.1 mM NH4+ compared to 0.5 to 1.0 mM NH4+. Environmental factors, such as pH and NH4+ availability, therefore need to be considered when using isotope effects in 15N isotope fractionation models of nitrification.
Increased microbial expression of organic nitrogen cycling genes in long-term warmed grassland soils
2021 - ISME Commun, 1: 69
Abstract:
Global warming increases soil temperatures and promotes faster growth and turnover of soil microbial communities. As microbial cell walls contain a high proportion of organic nitrogen, a higher turnover rate of microbes should also be reflected in an accelerated organic nitrogen cycling in soil. We used a metatranscriptomics and metagenomics approach to demonstrate that the relative transcription level of genes encoding enzymes involved in the extracellular depolymerization of high-molecular-weight organic nitrogen was higher in medium-term (8 years) and long-term (>50 years) warmed soils than in ambient soils. This was mainly driven by increased levels of transcripts coding for enzymes involved in the degradation of microbial cell walls and proteins. Additionally, higher transcription levels for chitin, nucleic acid, and peptidoglycan degrading enzymes were found in long-term warmed soils. We conclude that an acceleration in microbial turnover under warming is coupled to higher investments in N acquisition enzymes, particularly those involved in the breakdown and recycling of microbial residues, in comparison with ambient conditions.
Novel Alcaligenes ammonioxydans sp. nov. from wastewater treatment sludge oxidizes ammonia to N with a previously unknown pathway.
2021 - Environ Microbiol, 11: 6965-6980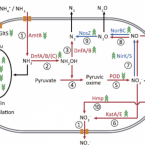
Abstract:
Heterotrophic nitrifiers are able to oxidize and remove ammonia from nitrogen-rich wastewaters but the genetic elements of heterotrophic ammonia oxidation are poorly understood. Here, we isolated and identified a novel heterotrophic nitrifier, Alcaligenes ammonioxydans sp. nov. strain HO-1, oxidizing ammonia to hydroxylamine and ending in the production of N gas. Genome analysis revealed that strain HO-1 encoded a complete denitrification pathway but lacks any genes coding for homologous to known ammonia monooxygenases or hydroxylamine oxidoreductases. Our results demonstrated strain HO-1 denitrified nitrite (not nitrate) to N and N O at anaerobic and aerobic conditions respectively. Further experiments demonstrated that inhibition of aerobic denitrification did not stop ammonia oxidation and N production. A gene cluster (dnfT1RT2ABCD) was cloned from strain HO-1 and enabled E. coli accumulated hydroxylamine. Sub-cloning showed that genetic cluster dnfAB or dnfABC already enabled E. coli cells to produce hydroxylamine and further to N from ( NH ) SO . Transcriptome analysis revealed these three genes dnfA, dnfB and dnfC were significantly upregulated in response to ammonia stimulation. Taken together, we concluded that strain HO-1 has a novel dnf genetic cluster for ammonia oxidation and this dnf genetic cluster encoded a previously unknown pathway of direct ammonia oxidation (Dirammox) to N .
Cyanate is a low abundance but actively cycled nitrogen compound in soil
2021 - Communications Earth & Environment, 2: 161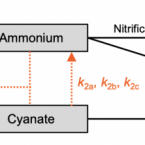
Abstract:
Cyanate can serve as a nitrogen and/or carbon source for different microorganisms and as an energy source for autotrophic ammonia oxidizers. However, the extent of cyanate availability and utilisation in terrestrial ecosystems and its role in biogeochemical cycles is poorly known. Here we analyse cyanate concentrations in soils across a range of soil types, land management practices and climates. Soil cyanate concentrations were three orders of magnitude lower than ammonium or nitrate. We determined cyanate consumption in a grassland and rice paddy soil using stable isotope tracer experiments. We find that cyanate turnover was rapid and dominated by biotic processes. We estimated that in-situ cyanate production rates were similar to those associated with urea fertilizer decomposition, a major source of cyanate in the environment. We provide evidence that cyanate is actively turned over in soils and represents a small but continuous nitrogen/energy source for soil microbes.
Sustained nitrogen loss in a symbiotic association of Comammox Nitrospira and Anammox bacteria
2021 - Water Res, 202: 117426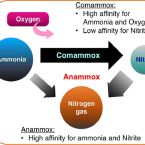
Abstract:
The discovery of anaerobic ammonia-oxidizing bacteria (Anammox) and, more recently, aerobic bacteria common in many natural and engineered systems that oxidize ammonia completely to nitrate (Comammox) have significantly altered our understanding of the global nitrogen cycle. A high affinity for ammonia (Km(app),NH3 ≈ 63nM) and oxygen place Comammox Nitrospira inopinata, the first described isolate, in the same trophic category as organisms such as some ammonia-oxidizing archaea. However, N. inopinata has a relatively low affinity for nitrite (Km,NO2 ≈ 449.2μM) suggesting it would be less competitive for nitrite than other nitrite-consuming aerobes and anaerobes. We examined the ecological relevance of the disparate substrate affinities by coupling it with the Anammox bacterium Candidatus Brocadia anammoxidans. Synthetic communities of the two were established in hydrogel granules in which Comammox grew in the aerobic outer layer to provide Anammox with nitrite in the inner anoxic core to form dinitrogen gas. This spatial organization was confirmed with FISH imaging, supporting a mutualistic or commensal relationship. The functional significance of interspecies spatial organization was informed by the hydrogel encapsulation format, broadening our limited understanding of the interplay between these two species. The resulting low nitrate formation and the competitiveness of Comammox over other aerobic ammonia- and nitrite-oxidizers sets this ecological cooperation apart and points to potential biotechnological applications. Since nitrate is an undesirable product of wastewater treatment effluents, the Comammox-Anammox symbiosis may be of economic and ecological importance to reduce nitrogen contamination of receiving waters.
Genomic insights into diverse bacterial taxa that degrade extracellular DNA in marine sediments
2021 - Nat Microbiol, 6: 885–898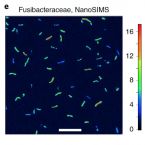
Abstract:
Extracellular DNA is a major macromolecule in global element cycles, and is a particularly crucial phosphorus, nitrogen and carbon source for microorganisms in the seafloor. Nevertheless, the identities, ecophysiology and genetic features of DNA-foraging microorganisms in marine sediments are largely unknown. Here, we combined microcosm experiments, DNA stable isotope probing (SIP), single-cell SIP using nano-scale secondary isotope mass spectrometry (NanoSIMS) and genome-centric metagenomics to study microbial catabolism of DNA and its subcomponents in marine sediments. 13C-DNA added to sediment microcosms was largely degraded within 10 d and mineralized to 13CO2. SIP probing of DNA revealed diverse ‘Candidatus Izemoplasma’, Lutibacter, Shewanella and Fusibacteraceae incorporated DNA-derived 13C-carbon. NanoSIMS confirmed incorporation of 13C into individual bacterial cells of Fusibacteraceae sorted from microcosms. Genomes of the 13C-labelled taxa all encoded enzymatic repertoires for catabolism of DNA or subcomponents of DNA. Comparative genomics indicated that diverse ‘Candidatus Izemoplasmatales’ (former Tenericutes) are exceptional because they encode multiple (up to five) predicted extracellular nucleases and are probably specialized DNA-degraders. Analyses of additional sediment metagenomes revealed extracellular nuclease genes are prevalent among Bacteroidota at diverse sites. Together, our results reveal the identities and functional properties of microorganisms that may contribute to the key ecosystem function of degrading and recycling DNA in the seabed.
Recently photoassimilated carbon and fungus-delivered nitrogen are spatially correlated in the ectomycorrhizal tissue of Fagus sylvatica.
2021 - New Phytol, 6: 2457-2474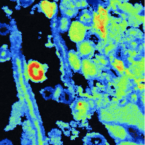
Abstract:
Ectomycorrhizal plants trade plant-assimilated carbon for soil nutrients with their fungal partners. The underlying mechanisms, however, are not fully understood. Here we investigate the exchange of carbon for nitrogen in the ectomycorrhizal symbiosis of Fagus sylvatica across different spatial scales from the root system to the cellular level. We provided N-labelled nitrogen to mycorrhizal hyphae associated with one half of the root system of young beech trees, while exposing plants to a CO atmosphere. We analysed the short-term distribution of C and N in the root system with isotope-ratio mass spectrometry, and at the cellular scale within a mycorrhizal root tip with nanoscale secondary ion mass spectrometry (NanoSIMS). At the root system scale, plants did not allocate more C to root parts that received more N. Nanoscale secondary ion mass spectrometry imaging, however, revealed a highly heterogenous, and spatially significantly correlated distribution of C and N at the cellular scale. Our results indicate that, on a coarse scale, plants do not allocate a larger proportion of photoassimilated C to root parts associated with N-delivering ectomycorrhizal fungi. Within the ectomycorrhizal tissue, however, recently plant-assimilated C and fungus-delivered N were spatially strongly coupled. Here, NanoSIMS visualisation provides an initial insight into the regulation of ectomycorrhizal C and N exchange at the microscale.
Novel taxa of Acidobacteriota implicated in seafloor sulfur cycling.
2021 - ISME J, 15: 3159–3180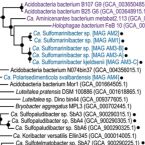
Abstract:
Acidobacteriota are widespread and often abundant in marine sediments, yet their metabolic and ecological properties are poorly understood. Here, we examined metabolisms and distributions of Acidobacteriota in marine sediments of Svalbard by functional predictions from metagenome-assembled genomes (MAGs), amplicon sequencing of 16S rRNA and dissimilatory sulfite reductase (dsrB) genes and transcripts, and gene expression analyses of tetrathionate-amended microcosms. Acidobacteriota were the second most abundant dsrB-harboring (averaging 13%) phylum after Desulfobacterota in Svalbard sediments, and represented 4% of dsrB transcripts on average. Meta-analysis of dsrAB datasets also showed Acidobacteriota dsrAB sequences are prominent in marine sediments worldwide, averaging 15% of all sequences analysed, and represent most of the previously unclassified dsrAB in marine sediments. We propose two new Acidobacteriota genera, Candidatus Sulfomarinibacter (class Thermoanaerobaculia, "subdivision 23") and Ca. Polarisedimenticola ("subdivision 22"), with distinct genetic properties that may explain their distributions in biogeochemically distinct sediments. Ca. Sulfomarinibacter encode flexible respiratory routes, with potential for oxygen, nitrous oxide, metal-oxide, tetrathionate, sulfur and sulfite/sulfate respiration, and possibly sulfur disproportionation. Potential nutrients and energy include cellulose, proteins, cyanophycin, hydrogen, and acetate. A Ca. Polarisedimenticola MAG encodes various enzymes to degrade proteins, and to reduce oxygen, nitrate, sulfur/polysulfide and metal-oxides. 16S rRNA gene and transcript profiling of Svalbard sediments showed Ca. Sulfomarinibacter members were relatively abundant and transcriptionally active in sulfidic fjord sediments, while Ca. Polarisedimenticola members were more relatively abundant in metal-rich fjord sediments. Overall, we reveal various physiological features of uncultured marine Acidobacteriota that indicate fundamental roles in seafloor biogeochemical cycling.
Electrochemical enrichment of marine denitrifying bacteria to enhance nitrate metabolization in seawater
2021 - J Environ Chem Eng, 9: 105604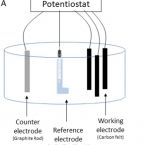
Abstract:
High concentrations of nitrate from industrial discharges to coastal marine environments are a matter of concern owing to their ecological consequences. In the last years, Bioelectrochemical Denitrification Systems (BEDS) have emerged as a promising nitrate removal technology. However, they still have limitations, such as the enrichment strategy for specific microbial communities in the electrodes under natural conditions. In this study, three-electrode electrochemical cells were used to test microbial enrichment from natural seawater by applying three reported potentials associated with the dissimilatory denitrification process (-130, -260, and -570 mV vs. Ag/AgCl). The microbial community analysis showed that by applying -260 mV (vs. Ag/AgCl) to the working electrode, it was possible to significantly enrich denitrifying microorganisms, specifically Marinobacter, in comparison with the control. Furthermore, -260 mV (vs. Ag/AgCl) led to a significantly higher nitrate removal than other conditions, which, combined with cyclic voltammetry analysis, suggested that the polarized electrodes worked as external electron donors for nitrate reduction. Hence, this work demonstrates for the first time that it is possible to enrich marine denitrifying microorganisms by applying an overpotential of -260 mV (vs. Ag/AgCl) without the need for a culture medium, the addition of an exogenous electron donor (i.e., organic matter) or a previously enriched inoculum.
Survival strategies of ammonia-oxidizing archaea (AOA) in a full-scale WWTP treating mixed landfill leachate containing copper ions and operating at low-intensity of aeration.
2021 - Water Res, 116798
Abstract:
Recent studies indicate that ammonia-oxidizing archaea (AOA) may play an important role in nitrogen removal by wastewater treatment plants (WWTPs). However, our knowledge of the mechanisms employed by AOA for growth and survival in full-scale WWTPs is still limited. Here, metagenomic and metatranscriptomic analyses combined with a laboratory cultivation experiment revealed that three active AOAs (WS9, WS192, and WS208) belonging to family Nitrososphaeraceae were active in the deep oxidation ditch (DOD) of a full-scale WWTP treating landfill leachate, which is configured with three continuous aerobic-anoxic (OA) modules with low-intensity aeration (≤ 1.5 mg/L). AOA coexisted with AOB and complete ammonia oxidizers (Comammox), while the ammonia-oxidizing microbial (AOM) community was unexpectedly dominated by the novel AOA strain WS9. The low aeration, long retention time, and relatively high inputs of ammonium and copper might be responsible for the survival of AOA over AOB and Comammox, while the dominance of WS9, specifically may be enhanced by substrate preference and uniquely encoded retention strategies. The urease-negative WS9 is specifically adapted for ammonia acquisition as evidenced by the high expression of an ammonium transporter, whereas two metabolically versatile urease-positive AOA strains (WS192 and WS208) can likely supplement ammonia needs with urea. This study provides important information for the survival and application of the eutrophic Nitrososphaeraceae AOA and advances our understanding of archaea-dominated ammonia oxidation in a full-scale wastewater treatment system.
Anaerobic bacterial degradation of protein and lipid macromolecules in subarctic marine sediment
2021 - ISME J, 15: 833-847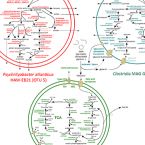
Abstract:
Microorganisms in marine sediments play major roles in marine biogeochemical cycles by mineralizing substantial quantities of organic matter from decaying cells. Proteins and lipids are abundant components of necromass, yet the taxonomic identities of microorganisms that actively degrade them remain poorly resolved. Here, we revealed identities, trophic interactions and genomic features of bacteria that degraded 13C-labelled proteins and lipids in cold anoxic microcosms containing sulfidic subarctic marine sediment. Supplemented proteins and lipids were rapidly fermented to various volatile fatty acids within five days. DNA-stable isotope probing (SIP) suggested Psychrilyobacter atlanticus was an important primary degrader of proteins, and Psychromonas members were important primary degraders of both proteins and lipids. Closely related Psychromonas populations, as represented by distinct 16S rRNA gene variants, differentially utilized either proteins or lipids. DNA-SIP also showed 13C-labeling of various Deltaproteobacteria within ten days, indicating trophic transfer of carbon to putative sulfate-reducers. Metagenome-assembled genomes revealed the primary hydrolyzers encoded secreted peptidases or lipases, and enzymes for catabolism of protein or lipid degradation products. Psychromonas species are prevalent in diverse marine sediments, suggesting they are important players in organic carbon processing in situ. Together, this study provides new insights into the identities, functions and genomes of bacteria that actively degrade abundant necromass macromolecules in the seafloor.
Genomic and kinetic analysis of novel Nitrospinae enriched by cell sorting.
2021 - ISME J, 15: 732–745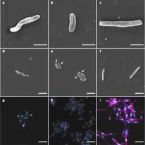
Abstract:
Chemolithoautotrophic nitrite-oxidizing bacteria (NOB) are key players in global nitrogen and carbon cycling. Members of the phylum Nitrospinae are the most abundant, known NOB in the oceans. To date, only two closely affiliated Nitrospinae species have been isolated, which are only distantly related to the environmentally abundant uncultured Nitrospinae clades. Here, we applied live cell sorting, activity screening, and subcultivation on marine nitrite-oxidizing enrichments to obtain novel marine Nitrospinae. Two binary cultures were obtained, each containing one Nitrospinae strain and one alphaproteobacterial heterotroph. The Nitrospinae strains represent two new genera, and one strain is more closely related to environmentally abundant Nitrospinae than previously cultured NOB. With an apparent half-saturation constant of 8.7 ± 2.5 µM, this strain has the highest affinity for nitrite among characterized marine NOB, while the other strain (16.2 ± 1.6 µM) and Nitrospina gracilis (20.1 ± 2.1 µM) displayed slightly lower nitrite affinities. The new strains and N. gracilis share core metabolic pathways for nitrite oxidation and CO fixation but differ remarkably in their genomic repertoires of terminal oxidases, use of organic N sources, alternative energy metabolisms, osmotic stress and phage defense. The new strains, tentatively named "Candidatus Nitrohelix vancouverensis" and "Candidatus Nitronauta litoralis", shed light on the niche differentiation and potential ecological roles of Nitrospinae.
Flow-through stable isotope probing (Flow-SIP) minimizes cross-feeding in complex microbial communities.
2021 - ISME J, 1: 348-353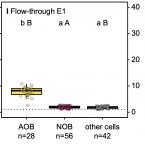
Abstract:
Stable isotope probing (SIP) is a key tool for identifying the microorganisms catalyzing the turnover of specific substrates in the environment and to quantify their relative contributions to biogeochemical processes. However, SIP-based studies are subject to the uncertainties posed by cross-feeding, where microorganisms release isotopically labeled products, which are then used by other microorganisms, instead of incorporating the added tracer directly. Here, we introduce a SIP approach that has the potential to strongly reduce cross-feeding in complex microbial communities. In this approach, the microbial cells are exposed on a membrane filter to a continuous flow of medium containing isotopically labeled substrate. Thereby, metabolites and degradation products are constantly removed, preventing consumption of these secondary substrates. A nanoSIMS-based proof-of-concept experiment using nitrifiers in activated sludge and C-bicarbonate as an activity tracer showed that Flow-SIP significantly reduces cross-feeding and thus allows distinguishing primary consumers from other members of microbial food webs.