Publications
Publications in peer reviewed journals
Metabolic and phylogenetic diversity in the phylum Nitrospinota revealed by comparative genome analyses
2024 - ISME Commun., 4: ycad017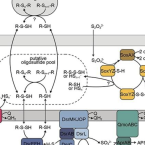
Abstract:
The most abundant known nitrite-oxidizing bacteria in the marine water column belong to the phylum Nitrospinota. Despite their importance in marine nitrogen cycling and primary production, there are only few cultured representatives that all belong to the class Nitrospinia. Moreover, although Nitrospinota were traditionally thought to be restricted to marine environments, metagenome-assembled genomes have also been recovered from groundwater. Over the recent years, metagenomic sequencing has led to the discovery of several novel classes of Nitrospinota (UBA9942, UBA7883, 2-12-FULL-45-22, JACRGO01, JADGAW01), which remain uncultivated and have not been analyzed in detail. Here, we analyzed a nonredundant set of 98 Nitrospinota genomes with focus on these understudied Nitrospinota classes and compared their metabolic profiles to get insights into their potential role in biogeochemical element cycling. Based on phylogenomic analysis and average amino acid identities, the highly diverse phylum Nitrospinota could be divided into at least 33 different genera, partly with quite distinct metabolic capacities. Our analysis shows that not all Nitrospinota are nitrite oxidizers and that members of this phylum have the genomic potential to use sulfide and hydrogen for energy conservation. This study expands our knowledge of the phylogeny and potential ecophysiology of the phylum Nitrospinota and offers new avenues for the isolation and cultivation of these elusive bacteria.
Cultivation and genomic characterization of novel and ubiquitous marine nitrite-oxidizing bacteria from the Nitrospirales
2023 - ISME J., in press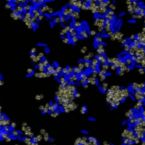
Abstract:
Nitrospirales, including the genus Nitrospira, are environmentally widespread chemolithoautotrophic nitrite-oxidizing bacteria. These mostly uncultured microorganisms gain energy through nitrite oxidation, fix CO2, and thus play vital roles in nitrogen and carbon cycling. Over the last decade, our understanding of their physiology has advanced through several new discoveries, such as alternative energy metabolisms and complete ammonia oxidizers (comammox Nitrospira). These findings mainly resulted from studies of terrestrial species, whereas less attention has been given to marine Nitrospirales. In this study, we cultured three new marine Nitrospirales enrichments and one isolate. Three of these four NOB represent new Nitrospira species while the fourth represents a novel genus. This fourth organism, tentatively named “Ca. Nitronereus thalassa”, represents the first cultured member of a Nitrospirales lineage that encompasses both free-living and sponge-associated nitrite oxidizers, is highly abundant in the environment, and shows distinct habitat distribution patterns compared to the marine Nitrospira species. Partially explaining this, “Ca. Nitronereus thalassa” harbors a unique combination of genes involved in carbon fixation and respiration, suggesting differential adaptations to fluctuating oxygen concentrations. Furthermore, “Ca. Nitronereus thalassa” appears to have a more narrow substrate range compared to many other marine nitrite oxidizers, as it lacks the genomic potential to utilize formate, cyanate, and urea. Lastly, we show that the presumed marine Nitrospirales lineages are not restricted to oceanic and saline environments, as previously assumed.
Nitrification in acidic and alkaline environments
2023 - Essays Biochem, in press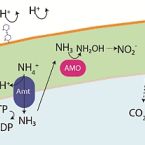
Abstract:
Aerobic nitrification is a key process in the global nitrogen cycle mediated by microorganisms. While nitrification has primarily been studied in near-neutral environments, this process occurs at a wide range of pH values, spanning ecosystems from acidic soils to soda lakes. Aerobic nitrification primarily occurs through the activities of ammonia-oxidising bacteria and archaea, nitrite-oxidising bacteria, and complete ammonia-oxidising (comammox) bacteria adapted to these environments. Here, we review the literature and identify knowledge gaps on the metabolic diversity, ecological distribution, and physiological adaptations of nitrifying microorganisms in acidic and alkaline environments. We emphasise that nitrifying microorganisms depend on a suite of physiological adaptations to maintain pH homeostasis, acquire energy and carbon sources, detoxify reactive nitrogen species, and generate a membrane potential at pH extremes. We also recognize the broader implications of their activities primarily in acidic environments, with a focus on agricultural productivity and nitrous oxide emissions, as well as promising applications in treating municipal wastewater.
Hot spring distribution and survival mechanisms of thermophilic comammox Nitrospira
2023 - ISME J., 17: 993-1003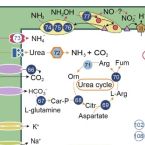
Abstract:
The recent discovery of Nitrospira species capable of complete ammonia oxidation (comammox) in non-marine natural and engineered ecosystems under mesothermal conditions has changed our understanding of microbial nitrification. However, little is known about the occurrence of comammox bacteria or their ability to survive in moderately thermal and/or hyperthermal habitats. Here, we report the wide distribution of comammox Nitrospira in five terrestrial hot springs at temperatures ranging from 36 to 80°C and provide metagenome-assembled genomes of 11 new comammox strains. Interestingly, the identification of dissimilatory nitrate reduction to ammonium (DNRA) in thermophilic comammox Nitrospira lineages suggests that they have versatile ecological functions as both sinks and sources of ammonia, in contrast to the described mesophilic comammox lineages, which lack the DNRA pathway. Furthermore, the in situ expression of key genes associated with nitrogen metabolism, thermal adaptation, and oxidative stress confirmed their ability to survive in the studied hot springs and their contribution to nitrification in these environments. Additionally, the smaller genome size and higher GC content, less polar and more charged amino acids in usage profiles, and the expression of a large number of heat shock proteins compared to mesophilic comammox strains presumably confer tolerance to thermal stress. These novel insights into the occurrence, metabolic activity, and adaptation of comammox Nitrospira in thermal habitats further expand our understanding of the global distribution of comammox Nitrospira and have significant implications for how these unique microorganisms have evolved thermal tolerance strategies.
Complete genome sequence of Nitrospina watsonii 347, isolated from the Black Sea
2023 - Microbiol Resour Announc, 12: e0007823
Abstract:
Here, we present the complete genome sequence of Nitrospina watsonii 347, a nitrite-oxidizing bacterium isolated from the Black Sea at a depth of 100 m. The genome has a length of 3,011,914 bp with 2,895 predicted coding sequences. Its predicted metabolism is similar to that of Nitrospina gracilis with differences in defense against reactive oxygen species.
Rapid nitrification involving comammox and canonical Nitrospira at extreme pH in saline-alkaline lakes.
2023 - Environ Microbiol, 25: 1055-1067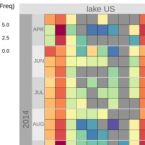
Abstract:
Nitrite-oxidizing bacteria (NOB) catalyse the second nitrification step and are the main biological source of nitrate. The most diverse and widespread NOB genus is Nitrospira, which also contains complete ammonia oxidizers (comammox) that oxidize ammonia to nitrate. To date, little is known about the occurrence and biology of comammox and canonical nitrite oxidizing Nitrospira in extremely alkaline environments. Here, we studied the seasonal distribution and diversity, and the effect of short-term pH changes on comammox and canonical Nitrospira in sediments of two saline, highly alkaline lakes. We identified diverse canonical and comammox Nitrospira clade A-like phylotypes as the only detectable NOB during more than a year, suggesting their major importance for nitrification in these habitats. Gross nitrification rates measured in microcosm incubations were highest at pH 10 and considerably faster than reported for other natural, aquatic environments. Nitrification could be attributed to canonical and comammox Nitrospira and to Nitrososphaerales ammonia-oxidizing archaea. Furthermore, our data suggested that comammox Nitrospira contributed to ammonia oxidation at an extremely alkaline pH of 11. These results identify saline, highly alkaline lake sediments as environments of uniquely strong nitrification with novel comammox Nitrospira as key microbial players.
Enhanced nitrogen and carbon removal in natural seawater by electrochemical enrichment in a bioelectrochemical reactor
2022 - J Environ Manage, 323: 116294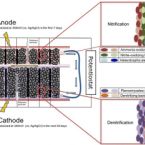
Abstract:
Municipal and industrial wastewater discharges in coastal and marine environments are of major concern due to their high carbon and nitrogen loads and the resulted phenomenon of eutrophication. Bioelectrochemical reactors (BERs) for simultaneous nitrogen and carbon removal have gained attention owing to their cost efficiency and versatility, as well as the possibility of electrochemical enrich specific groups. This study presented a scalable two-chamber BERs using graphite granules as electrode material. BERs were inoculated and operated for 37 days using natural seawater with high concentrations of ammonium and acetate. The BERs demonstrated a maximum current density of 0.9 A m−3 and removal rates of 7.5 mg NH4+-N L−1 d−1 and 99.5 mg L−1 d−1 for total organic carbon (TOC). Removals observed for NH4+-N and TOC were 96.2% and 68.7%, respectively. The results of nutrient removal (i.e., ammonium, nitrate, nitrite and TOC) and microbial characterization (i.e., next-generation sequencing of the 16S rRNA gene and fluorescence in situ hybridization) showed that BERs operated with a poised cathode at −260 mV (vs. Ag/AgCl) significantly enriched nitrifying microorganisms in the anode and denitrifying microorganisms and planctomycetes in the cathode. Interestingly, the electrochemical enrichment did not increase the total number of microorganisms in the formed biofilms but controlled their composition. Thus, this work shows the first successful attempt to electrochemically enrich marine nitrifying and denitrifying microorganisms and presents a technique to accelerate the start-up process of BERs to remove dissolved inorganic nitrogen and total organic carbon from seawater.
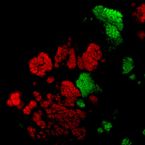
Specific localization and quantification of the Oligo-Mouse-Microbiota (OMM12) by fluorescence in situ hybridization (FISH)
2022 - Current Protocols, 2: e548
Abstract:
The oligo-mouse-microbiota (OMM12) is a widely used syncom that colonizes gnotobiotic mice in a stable manner. It provides several fundamental functions to its murine host, including colonization resistance against enteric pathogens. Here, we designed and validated specific fluorescence in situ hybridization (FISH) probes to detect and quantify OMM12 strains on intestinal tissue cross sections. 16S rRNA‒specific probes were designed, and specificity was validated on fixed pure cultures. A hybridization protocol was optimized for sensitive detection of the individual bacterial cells in cryosections. Using this method, we showed that the intestinal mucosal niche of Akkermansia muciniphila can be influenced by global gut microbial community context.
A nitrite-oxidising bacterium constitutively consumes atmospheric hydrogen
2022 - ISME J, 16: 2213-2219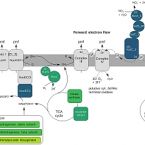
Abstract:
Chemolithoautotrophic nitrite-oxidising bacteria (NOB) of the genus Nitrospira contribute to nitrification in diverse natural environments and engineered systems. Nitrospira are thought to be well-adapted to substrate limitation owing to their high affinity for nitrite and capacity to use alternative energy sources. Here, we demonstrate that the canonical nitrite oxidiser Nitrospira moscoviensis oxidises hydrogen (H2) below atmospheric levels using a high-affinity group 2a nickel-iron hydrogenase [Km(app) = 32 nM]. Atmospheric H2 oxidation occurred under both nitrite-replete and nitrite-deplete conditions, suggesting low-potential electrons derived from H2 oxidation promote nitrite-dependent growth and enable survival during nitrite limitation. Proteomic analyses confirmed the hydrogenase was abundant under both conditions and indicated extensive metabolic changes occur to reduce energy expenditure and growth under nitrite-deplete conditions. Thermodynamic modelling revealed that H2 oxidation theoretically generates higher power yield than nitrite oxidation at low substrate concentrations and significantly contributes to growth at elevated nitrite concentrations. Collectively, this study suggests atmospheric H2 oxidation enhances the growth and survival of NOB amid variability of nitrite supply, extends the phenomenon of atmospheric H2 oxidation to an eighth phylum (Nitrospirota), and reveals unexpected new links between the global hydrogen and nitrogen cycles. Long classified as obligate nitrite oxidisers, our findings suggest H2 may primarily support growth and survival of certain NOB in natural environments.
Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities.
2022 - ISME J, 16: 272-283
Abstract:
Nitrification, the oxidation of ammonia to nitrate, is an essential process in the biogeochemical nitrogen cycle. The first step of nitrification, ammonia oxidation, is performed by three, often co-occurring guilds of chemolithoautotrophs: ammonia-oxidizing bacteria (AOB), archaea (AOA), and complete ammonia oxidizers (comammox). Substrate kinetics are considered to be a major niche-differentiating factor between these guilds, but few AOA strains have been kinetically characterized. Here, the ammonia oxidation kinetic properties of 12 AOA representing all major cultivated phylogenetic lineages were determined using microrespirometry. Members of the genus Nitrosocosmicus have the lowest affinity for both ammonia and total ammonium of any characterized AOA, and these values are similar to previously determined ammonia and total ammonium affinities of AOB. This contrasts previous assumptions that all AOA possess much higher substrate affinities than their comammox or AOB counterparts. The substrate affinity of ammonia oxidizers correlated with their cell surface area to volume ratios. In addition, kinetic measurements across a range of pH values supports the hypothesis that-like for AOB-ammonia and not ammonium is the substrate for the ammonia monooxygenase enzyme of AOA and comammox. Together, these data will facilitate predictions and interpretation of ammonia oxidizer community structures and provide a robust basis for establishing testable hypotheses on competition between AOB, AOA, and comammox.
Nitrogen kinetic isotope effects of nitrification by the complete ammonia oxidizer Nitrospira inopinata
2021 - mSphere, 6: e00634-21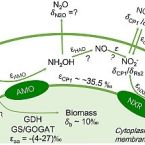
Abstract:
Analysis of nitrogen isotope fractionation effects is useful for tracing biogeochemical nitrogen cycle processes. Nitrification can cause large nitrogen isotope effects through the enzymatic oxidation of ammonia (NH3) via nitrite (NO2-) to nitrate (NO3-) (15εNH4+→NO2- and 15ɛNO2-→NO3-). The isotope effects of ammonia-oxidizing bacteria (AOB) and archaea (AOA), and nitrite-oxidizing bacteria (NOB), have been analyzed previously. Here we studied the nitrogen isotope effects of the complete ammonia oxidizer (comammox) Nitrospira inopinata that oxidizes NH3 to NO3-. At high ammonium (NH4+) availability (1 mM) and pH between 6.5 and 8.5, its 15εNH4+→NO2- ranged from −33.1 to −27.1‰ based on substrate consumption (residual substrate isotopic composition) and −35.5 to −31.2‰ based on product formation (cumulative product isotopic composition), while the 15ɛNO2-→NO3- ranged from 6.5 to 11.1‰ based on substrate consumption. These values resemble isotope effects of AOB and AOA, and of NOB in the genus Nitrospira, suggesting the absence of fundamental mechanistic differences between key enzymes for ammonia and nitrite oxidation in comammox and canonical nitrifiers. However, ambient pH and initial NH4+ concentrations influenced the isotope effects in N. inopinata. The 15εNH4+→NO2- based on product formation was smaller at pH 6.5 (−31.2‰) compared to pH 7.5 (−35.5‰) and pH 8.5 (−34.9‰), while 15ɛNO2-→NO3- was smaller at pH 8.5 (6.5‰) compared to pH 7.5 (8.8‰) and pH 6.5 (11.1‰). Isotopic fractionation via 15εNH4+→NO2- and 15ɛNO2-→NO3- was smaller at 0.1 mM NH4+ compared to 0.5 to 1.0 mM NH4+. Environmental factors, such as pH and NH4+ availability, therefore need to be considered when using isotope effects in 15N isotope fractionation models of nitrification.
Sustained nitrogen loss in a symbiotic association of Comammox Nitrospira and Anammox bacteria
2021 - Water Res, 202: 117426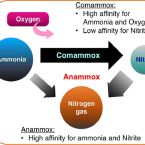
Abstract:
The discovery of anaerobic ammonia-oxidizing bacteria (Anammox) and, more recently, aerobic bacteria common in many natural and engineered systems that oxidize ammonia completely to nitrate (Comammox) have significantly altered our understanding of the global nitrogen cycle. A high affinity for ammonia (Km(app),NH3 ≈ 63nM) and oxygen place Comammox Nitrospira inopinata, the first described isolate, in the same trophic category as organisms such as some ammonia-oxidizing archaea. However, N. inopinata has a relatively low affinity for nitrite (Km,NO2 ≈ 449.2μM) suggesting it would be less competitive for nitrite than other nitrite-consuming aerobes and anaerobes. We examined the ecological relevance of the disparate substrate affinities by coupling it with the Anammox bacterium Candidatus Brocadia anammoxidans. Synthetic communities of the two were established in hydrogel granules in which Comammox grew in the aerobic outer layer to provide Anammox with nitrite in the inner anoxic core to form dinitrogen gas. This spatial organization was confirmed with FISH imaging, supporting a mutualistic or commensal relationship. The functional significance of interspecies spatial organization was informed by the hydrogel encapsulation format, broadening our limited understanding of the interplay between these two species. The resulting low nitrate formation and the competitiveness of Comammox over other aerobic ammonia- and nitrite-oxidizers sets this ecological cooperation apart and points to potential biotechnological applications. Since nitrate is an undesirable product of wastewater treatment effluents, the Comammox-Anammox symbiosis may be of economic and ecological importance to reduce nitrogen contamination of receiving waters.
Electrochemical enrichment of marine denitrifying bacteria to enhance nitrate metabolization in seawater
2021 - J Environ Chem Eng, 9: 105604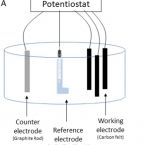
Abstract:
High concentrations of nitrate from industrial discharges to coastal marine environments are a matter of concern owing to their ecological consequences. In the last years, Bioelectrochemical Denitrification Systems (BEDS) have emerged as a promising nitrate removal technology. However, they still have limitations, such as the enrichment strategy for specific microbial communities in the electrodes under natural conditions. In this study, three-electrode electrochemical cells were used to test microbial enrichment from natural seawater by applying three reported potentials associated with the dissimilatory denitrification process (-130, -260, and -570 mV vs. Ag/AgCl). The microbial community analysis showed that by applying -260 mV (vs. Ag/AgCl) to the working electrode, it was possible to significantly enrich denitrifying microorganisms, specifically Marinobacter, in comparison with the control. Furthermore, -260 mV (vs. Ag/AgCl) led to a significantly higher nitrate removal than other conditions, which, combined with cyclic voltammetry analysis, suggested that the polarized electrodes worked as external electron donors for nitrate reduction. Hence, this work demonstrates for the first time that it is possible to enrich marine denitrifying microorganisms by applying an overpotential of -260 mV (vs. Ag/AgCl) without the need for a culture medium, the addition of an exogenous electron donor (i.e., organic matter) or a previously enriched inoculum.
Genomic and kinetic analysis of novel Nitrospinae enriched by cell sorting.
2021 - ISME J, 15: 732–745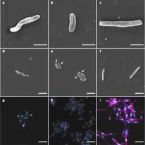
Abstract:
Chemolithoautotrophic nitrite-oxidizing bacteria (NOB) are key players in global nitrogen and carbon cycling. Members of the phylum Nitrospinae are the most abundant, known NOB in the oceans. To date, only two closely affiliated Nitrospinae species have been isolated, which are only distantly related to the environmentally abundant uncultured Nitrospinae clades. Here, we applied live cell sorting, activity screening, and subcultivation on marine nitrite-oxidizing enrichments to obtain novel marine Nitrospinae. Two binary cultures were obtained, each containing one Nitrospinae strain and one alphaproteobacterial heterotroph. The Nitrospinae strains represent two new genera, and one strain is more closely related to environmentally abundant Nitrospinae than previously cultured NOB. With an apparent half-saturation constant of 8.7 ± 2.5 µM, this strain has the highest affinity for nitrite among characterized marine NOB, while the other strain (16.2 ± 1.6 µM) and Nitrospina gracilis (20.1 ± 2.1 µM) displayed slightly lower nitrite affinities. The new strains and N. gracilis share core metabolic pathways for nitrite oxidation and CO fixation but differ remarkably in their genomic repertoires of terminal oxidases, use of organic N sources, alternative energy metabolisms, osmotic stress and phage defense. The new strains, tentatively named "Candidatus Nitrohelix vancouverensis" and "Candidatus Nitronauta litoralis", shed light on the niche differentiation and potential ecological roles of Nitrospinae.
A refined set of rRNA-targeted oligonucleotide probes for in situ detection and quantification of ammonia-oxidizing bacteria
2020 - Water Res., 186: 116372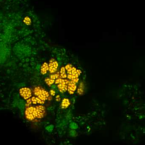
Abstract:
Ammonia-oxidizing bacteria (AOB) of the betaproteobacterial genera Nitrosomonas and Nitrosospira are key nitrifying microorganisms in many natural and engineered ecosystems. Since many AOB remain uncultured, fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes has been one of the most widely used approaches to study the community composition, abundance, and other features of AOB directly in environmental samples. However, the established and widely used AOB-specific 16S rRNA-targeted FISH probes were designed up to two decades ago, based on much smaller rRNA gene sequence datasets than available today. Several of these probes cover their target AOB lineages incompletely and suffer from a weak target specificity, which causes cross-hybridization of probes that should detect different AOB lineages. Here, a set of new highly specific 16S rRNA-targeted oligonucleotide probes was developed and experimentally evaluated that complements the existing probes and enables the specific detection and differentiation of the known, major phylogenetic clusters of betaproteobacterial AOB. The new probes were successfully applied to visualize and quantify AOB in activated sludge and biofilm samples from seven pilot- and full-scale wastewater treatment systems. Based on its improved target group coverage and specificity, the refined probe set will facilitate future in situ analyses of AOB.
Exploring the upper pH limits of nitrite oxidation: diversity, ecophysiology, and adaptive traits of haloalkalitolerant Nitrospira.
2020 - ISME J, 12: 2967-2979
Abstract:
Nitrite-oxidizing bacteria of the genus Nitrospira are key players of the biogeochemical nitrogen cycle. However, little is known about their occurrence and survival strategies in extreme pH environments. Here, we report on the discovery of physiologically versatile, haloalkalitolerant Nitrospira that drive nitrite oxidation at exceptionally high pH. Nitrospira distribution, diversity, and ecophysiology were studied in hypo- and subsaline (1.3-12.8 g salt/l), highly alkaline (pH 8.9-10.3) lakes by amplicon sequencing, metagenomics, and cultivation-based approaches. Surprisingly, not only were Nitrospira populations detected, but they were also considerably diverse with presence of members from Nitrospira lineages I, II and IV. Furthermore, the ability of Nitrospira enrichment cultures to oxidize nitrite at neutral to highly alkaline pH of 10.5 was demonstrated. Metagenomic analysis of a newly enriched Nitrospira lineage IV species, "Candidatus Nitrospira alkalitolerans", revealed numerous adaptive features of this organism to its extreme environment. Among them were a sodium-dependent N-type ATPase and NADH:quinone oxidoreductase next to the proton-driven forms usually found in Nitrospira. Other functions aid in pH and cation homeostasis and osmotic stress defense. "Ca. Nitrospira alkalitolerans" also possesses group 2a and 3b [NiFe] hydrogenases, suggesting it can use hydrogen as alternative energy source. These results reveal how Nitrospira cope with strongly fluctuating pH and salinity conditions and expand our knowledge of nitrogen cycling in extreme habitats.
Activity and metabolic versatility of complete ammonia oxidizers in full-scale wastewater treatment systems.
2020 - mBio, 11: e03175-19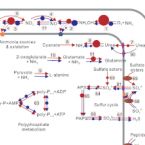
Abstract:
The recent discovery of complete ammonia oxidizers (comammox) contradicts the paradigm that chemolithoautotrophic nitrification is always catalyzed by two different microorganisms. However, our knowledge of the survival strategies of comammox in complex ecosystems, such as full-scale wastewater treatment plants (WWTPs), remains limited. Analyses of genomes and transcriptomes of four comammox organisms from two full-scale WWTPs revealed that comammox were active and showed a surprisingly high metabolic versatility. A gene cluster for the utilization of urea and a gene encoding cyanase suggest that comammox may use diverse organic nitrogen compounds in addition to free ammonia as the substrates. The comammox organisms also encoded the genomic potential for multiple alternative energy metabolisms, including respiration with hydrogen, formate, and sulfite as electron donors. Pathways for the biosynthesis and degradation of polyphosphate, glycogen, and polyhydroxyalkanoates as intracellular storage compounds likely help comammox survive unfavorable conditions and facilitate switches between lifestyles in fluctuating environments. One of the comammox strains acquired from the anaerobic tank encoded and transcribed genes involved in homoacetate fermentation or in the utilization of exogenous acetate, both pathways being unexpected in a nitrifying bacterium. Surprisingly, this strain also encoded a respiratory nitrate reductase which has not yet been found in any other genome and might confer a selective advantage to this strain over other strains in anoxic conditions. The discovery of comammox in the genus changes our perception of nitrification. However, genomes of comammox organisms have not been acquired from full-scale WWTPs, and very little is known about their survival strategies and potential metabolisms in complex wastewater treatment systems. Here, four comammox metagenome-assembled genomes and metatranscriptomic data sets were retrieved from two full-scale WWTPs. Their impressive and-among nitrifiers-unsurpassed ecophysiological versatility could make comammox an interesting target for optimizing nitrification in current and future bioreactor configurations.
Single cell analyses reveal contrasting life strategies of the two main nitrifiers in the ocean.
2020 - Nat Commun, 1: 767
Abstract:
Nitrification, the oxidation of ammonia via nitrite to nitrate, is a key process in marine nitrogen (N) cycling. Although oceanic ammonia and nitrite oxidation are balanced, ammonia-oxidizing archaea (AOA) vastly outnumber the main nitrite oxidizers, the bacterial Nitrospinae. The ecophysiological reasons for this discrepancy in abundance are unclear. Here, we compare substrate utilization and growth of Nitrospinae to AOA in the Gulf of Mexico. Based on our results, more than half of the Nitrospinae cellular N-demand is met by the organic-N compounds urea and cyanate, while AOA mainly assimilate ammonium. Nitrospinae have, under in situ conditions, around four-times higher biomass yield and five-times higher growth rates than AOA, despite their ten-fold lower abundance. Our combined results indicate that differences in mortality between Nitrospinae and AOA, rather than thermodynamics, biomass yield and cell size, determine the abundances of these main marine nitrifiers. Furthermore, there is no need to invoke yet undiscovered, abundant nitrite oxidizers to explain nitrification rates in the ocean.
Transcriptomic Response of Nitrosomonas europaea Transitioned from Ammonia- to Oxygen-Limited Steady-State Growth.
2020 - mSystems, 1: e00562-19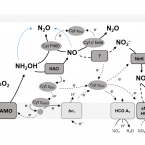
Abstract:
Ammonia-oxidizing microorganisms perform the first step of nitrification, the oxidation of ammonia to nitrite. The bacterium is the best-characterized ammonia oxidizer to date. Exposure to hypoxic conditions has a profound effect on the physiology of , e.g., by inducing nitrifier denitrification, resulting in increased nitric and nitrous oxide production. This metabolic shift is of major significance in agricultural soils, as it contributes to fertilizer loss and global climate change. Previous studies investigating the effect of oxygen limitation on have focused on the transcriptional regulation of genes involved in nitrification and nitrifier denitrification. Here, we combine steady-state cultivation with whole-genome transcriptomics to investigate the overall effect of oxygen limitation on Under oxygen-limited conditions, growth yield was reduced and ammonia-to-nitrite conversion was not stoichiometric, suggesting the production of nitrogenous gases. However, the transcription of the principal nitric oxide reductase (cNOR) did not change significantly during oxygen-limited growth, while the transcription of the nitrite reductase-encoding gene () was significantly lower. In contrast, both heme-copper-containing cytochrome oxidases encoded by were upregulated during oxygen-limited growth. Particularly striking was the significant increase in transcription of the B-type heme-copper oxidase, proposed to function as a nitric oxide reductase (sNOR) in ammonia-oxidizing bacteria. In the context of previous physiological studies, as well as the evolutionary placement of sNOR with regard to other heme-copper oxidases, these results suggest sNOR may function as a high-affinity terminal oxidase in and other ammonia-oxidizing bacteria. Nitrification is a ubiquitous microbially mediated process in the environment and an essential process in engineered systems such as wastewater and drinking water treatment plants. However, nitrification also contributes to fertilizer loss from agricultural environments, increasing the eutrophication of downstream aquatic ecosystems, and produces the greenhouse gas nitrous oxide. As ammonia-oxidizing bacteria are the most dominant ammonia-oxidizing microbes in fertilized agricultural soils, understanding their responses to a variety of environmental conditions is essential for curbing the negative environmental effects of nitrification. Notably, oxygen limitation has been reported to significantly increase nitric oxide and nitrous oxide production during nitrification. Here, we investigate the physiology of the best-characterized ammonia-oxidizing bacterium, , growing under oxygen-limited conditions.
A fiber-deprived diet disturbs the fine-scale spatial architecture of the murine colon microbiome.
2019 - Nat Commun, 1: 4366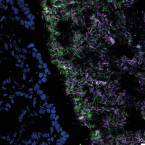
Abstract:
Compartmentalization of the gut microbiota is thought to be important to system function, but the extent of spatial organization in the gut ecosystem remains poorly understood. Here, we profile the murine colonic microbiota along longitudinal and lateral axes using laser capture microdissection. We found fine-scale spatial structuring of the microbiota marked by gradients in composition and diversity along the length of the colon. Privation of fiber reduces the diversity of the microbiota and disrupts longitudinal and lateral gradients in microbiota composition. Both mucus-adjacent and luminal communities are influenced by the absence of dietary fiber, with the loss of a characteristic distal colon microbiota and a reduction in the mucosa-adjacent community, concomitant with depletion of the mucus layer. These results indicate that diet has not only global but also local effects on the composition of the gut microbiota, which may affect function and resilience differently depending on location.
A Multicolor Fluorescence Hybridization Approach Using an Extended Set of Fluorophores to Visualize Microorganisms.
2019 - Front Microbiol, 10: 1383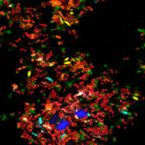
Abstract:
Fluorescence hybridization (FISH) with rRNA-targeted oligonucleotide probes is a key method for the detection of (uncultured) microorganisms in environmental and medical samples. A major limitation of standard FISH protocols, however, is the small number of phylogenetically distinct target organisms that can be detected simultaneously. In this study, we introduce a multicolor FISH approach that uses eight fluorophores with distinct spectral properties, which can unambiguously be distinguished by confocal laser scanning microscopy combined with white light laser technology. Hybridization of rRNA-targeted DNA oligonucleotide probes, which were mono-labeled with these fluorophores, to cultures confirmed that the fluorophores did not affect probe melting behavior. Application of the new multicolor FISH method enabled the differentiation of seven (potentially up to eight) phylogenetically distinct microbial populations in an artificial community of mixed pure cultures (five bacteria, one archaeon, and one yeast strain) and in activated sludge from a full-scale wastewater treatment plant. In contrast to previously published multicolor FISH approaches, this method does not rely on combinatorial labeling of the same microorganisms with different fluorophores, which is prone to biases. Furthermore, images acquired by this method do not require elaborate post-processing prior to analysis. We also demonstrate that the newly developed multicolor FISH method is compatible with an improved cell fixation protocol for FISH targeting Gram-negative bacterial populations. This fixation approach uses agarose embedding during formaldehyde fixation to better preserve the three-dimensional structure of spatially complex samples such as biofilms and activated sludge flocs. The new multicolor FISH approach should be highly suitable for studying structural and functional aspects of microbial communities in virtually all types of samples that can be analyzed by conventional FISH methods.
Low yield and abiotic origin of N2O formed by the complete nitrifier Nitrospira inopinata.
2019 - Nat Commun, 1: 1836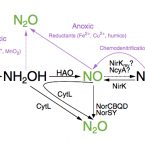
Abstract:
Nitrous oxide (NO) and nitric oxide (NO) are atmospheric trace gases that contribute to climate change and affect stratospheric and ground-level ozone concentrations. Ammonia oxidizing bacteria (AOB) and archaea (AOA) are key players in the nitrogen cycle and major producers of NO and NO globally. However, nothing is known about NO and NO production by the recently discovered and widely distributed complete ammonia oxidizers (comammox). Here, we show that the comammox bacterium Nitrospira inopinata is sensitive to inhibition by an NO scavenger, cannot denitrify to NO, and emits NO at levels that are comparable to AOA but much lower than AOB. Furthermore, we demonstrate that NO formed by N. inopinata formed under varying oxygen regimes originates from abiotic conversion of hydroxylamine. Our findings indicate that comammox microbes may produce less NO during nitrification than AOB.
An automated Raman-based platform for the sorting of live cells by functional properties.
2019 - Nat Microbiol, 6: 1035-1048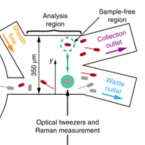
Abstract:
Stable-isotope probing is widely used to study the function of microbial taxa in their natural environment, but sorting of isotopically labelled microbial cells from complex samples for subsequent genomic analysis or cultivation is still in its early infancy. Here, we introduce an optofluidic platform for automated sorting of stable-isotope-probing-labelled microbial cells, combining microfluidics, optical tweezing and Raman microspectroscopy, which yields live cells suitable for subsequent single-cell genomics, mini-metagenomics or cultivation. We describe the design and optimization of this Raman-activated cell-sorting approach, illustrate its operation with four model bacteria (two intestinal, one soil and one marine) and demonstrate its high sorting accuracy (98.3 ± 1.7%), throughput (200-500 cells h; 3.3-8.3 cells min) and compatibility with cultivation. Application of this sorting approach for the metagenomic characterization of bacteria involved in mucin degradation in the mouse colon revealed a diverse consortium of bacteria, including several members of the underexplored family Muribaculaceae, highlighting both the complexity of this niche and the potential of Raman-activated cell sorting for identifying key players in targeted processes.
The draft genome sequence of “Nitrospira lenta” strain BS10, a nitrite oxidizing bacterium isolated from activated sludge
2018 - Stand Genomic Sci, 13: 32
Abstract:
The genus Nitrospira is considered to be the most widespread and abundant group of nitrite-oxidizing bacteria in many natural and man-made ecosystems. However, the ecophysiological versatility within this phylogenetic group remains highly understudied, mainly due to the lack of pure cultures and genomic data. To further expand our understanding of this biotechnologically important genus, we analyzed the high quality draft genome of “Nitrospira lenta” strain BS10, a sublineage II Nitrospira that was isolated from a municipal wastewater treatment plant in Hamburg, Germany. The genome of “N. lenta” has a size of 3,756,190 bp and contains 3968 genomic objects, of which 3907 are predicted protein-coding sequences. Thorough genome annotation allowed the reconstruction of the “N. lenta” core metabolism for energy conservation and carbon fixation. Comparative analyses indicated that most metabolic features are shared with N. moscoviensis and “N. defluvii”, despite their ecological niche differentiation and phylogenetic distance. In conclusion, the genome of “N. lenta” provides important insights into the genomic diversity of the genus Nitrospira and provides a foundation for future comparative genomic studies that will generate a better understanding of the nitrification process.
Characterization of the first “Candidatus Nitrotoga” isolate reveals metabolic versatility and separate evolution of widespread nitrite-oxidizing bacteria
2018 - mBio, 9: e01186-18
Abstract:
Nitrification is a key process of the biogeochemical nitrogen cycle and of biological wastewater treatment. The second step, nitrite oxidation to nitrate, is catalyzed by phylogenetically diverse, chemolithoautotrophic nitrite-oxidizing bacteria (NOB). Uncultured NOB from the genus “Candidatus Nitrotoga” are widespread in natural and engineered ecosystems. Knowledge about their biology is sparse, because no genomic information and no pure “Ca. Nitrotoga” culture was available. Here we obtained the first “Ca. Nitrotoga” isolate from activated sludge. This organism, “Candidatus Nitrotoga fabula,” prefers higher temperatures (>20°C; optimum, 24 to 28°C) than previous “Ca. Nitrotoga” enrichments, which were described as cold-adapted NOB. “Ca. Nitrotoga fabula” also showed an unusually high tolerance to nitrite (activity at 30 mM NO2−) and nitrate (up to 25 mM NO3−). Nitrite oxidation followed Michaelis-Menten kinetics, with an apparent Km (Km(app)) of ~89 µM nitrite and a Vmax of ~28 µmol of nitrite per mg of protein per h. Key metabolic pathways of “Ca. Nitrotoga fabula” were reconstructed from the closed genome. “Ca. Nitrotoga fabula” possesses a new type of periplasmic nitrite oxidoreductase belonging to a lineage of mostly uncharacterized proteins. This novel enzyme indicates (i) separate evolution of nitrite oxidation in “Ca. Nitrotoga” and other NOB, (ii) the possible existence of phylogenetically diverse, unrecognized NOB, and (iii) together with new metagenomic data, the potential existence of nitrite-oxidizing archaea. For carbon fixation, “Ca. Nitrotoga fabula” uses the Calvin-Benson-Bassham cycle. It also carries genes encoding complete pathways for hydrogen and sulfite oxidation, suggesting that alternative energy metabolisms enable “Ca. Nitrotoga fabula” to survive nitrite depletion and colonize new niches.
Cultivation and genomic analysis of “Candidatus Nitrosocaldus islandicus”, an obligately thermophilic, ammonia-oxidizing thaumarchaeon from a hot spring biofilm in Graendalur valley, Iceland
2018 - Front Microbiol, 9: 193
Abstract:
Ammonia-oxidizing archaea (AOA) within the phylum Thaumarchaeota are the only known aerobic ammonia oxidizers in geothermal environments. Although molecular data indicate the presence of phylogenetically diverse AOA from the Nitrosocaldus clade, group 1.1b and group 1.1a Thaumarchaeota in terrestrial high-temperature habitats, only one enrichment culture of an AOA thriving above 50 °C has been reported and functionally analyzed. In this study, we physiologically and genomically characterized a newly discovered thaumarchaeon from the deep-branching Nitrosocaldaceae family of which we have obtained a high (~85 %) enrichment from biofilm of an Icelandic hot spring (73 °C). This AOA, which we provisionally refer to as “Candidatus Nitrosocaldus islandicus”, is an obligately thermophilic, aerobic chemolithoautotrophic ammonia oxidizer, which stoichiometricall converts ammonia to nitrite at temperatures between 50 °C and 70 °C. “Ca. N. islandicus” encodes the expected repertoire of enzymes proposed to be required for archaeal ammonia oxidation, but unexpectedly lacks a nirK gene and also possesses no identifiable other enzyme for nitric oxide (NO) generation*. Nevertheless, ammonia oxidation by this AOA appears to be NO-dependent as “Ca. N. islandicus” is, like all other tested AOA, inhibited by the addition of an NO scavenger. Furthermore, comparative genomics revealed that “Ca. N. islandicus” has the potential for aromatic amino acid fermentation as its genome encodes an indolepyruvate oxidoreductase (iorAB) as well as a type 3b hydrogenase, which are not present in any other sequenced AOA. A further surprising genomic feature of this thermophilic ammonia oxidizer is the absence of DNA polymerase D genes – one of the predominant replicative DNA polymerases in all other ammonia-oxidizing Thaumarchaeota. Collectively, our findings suggest that metabolic versatility and DNA replication might differ substantially between obligately thermophilic and other AOA.
Draft genome sequence of Telmatospirillum siberiense 26-4b1T, an acidotolerant peatland alphaproteobacterium potentially involved in sulfur cycling
2018 - Genome Announc, 6: e01524-17
Abstract:
The facultative anaerobic chemoorganoheterotrophic alphaproteobacterium Telmatospirillum siberiense 26-4b1T was isolated from a Siberian peatland. We report on a 6.20 Mbp near complete, high quality draft genome of T. siberiense that reveals expected and novel metabolic potential for the genus Telmatospirillum, including genes for sulfur oxidation.
Adaptability as the key to success for the ubiquitous marine nitrite oxidizer Nitrococcus
2017 - Sci Adv, 3: e1700807
Abstract:
Nitrite-oxidizing bacteria (NOB) have conventionally been regarded as a highly specialized functional group responsible for the production of nitrate in the environment. However, recent culture-based studies suggest that they have the capacity to lead alternative lifestyles, but direct environmental evidence for the contribution of marine nitrite oxidizers to other processes has been lacking to date. We report on the alternative biogeochemical functions, worldwide distribution, and sometimes high abundance of the marine NOB Nitrococcus. These largely overlooked bacteria are capable of not only oxidizing nitrite but also reducing nitrate and producing nitrous oxide, an ozone-depleting agent and greenhouse gas. Furthermore, Nitrococcus can aerobically oxidize sulfide, thereby also engaging in the sulfur cycle. In the currently fast-changing global oceans, these findings highlight the potential functional switches these ubiquitous bacteria can perform in various biogeochemical cycles, each with distinct or even contrasting consequences.
Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle.
2017 - Nature, 549: 269-272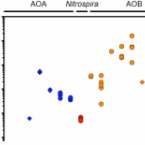
Abstract:
Nitrification, the oxidation of ammonia (NH3) via nitrite (NO2(-)) to nitrate (NO3(-)), is a key process of the biogeochemical nitrogen cycle. For decades, ammonia and nitrite oxidation were thought to be separately catalysed by ammonia-oxidizing bacteria (AOB) and archaea (AOA), and by nitrite-oxidizing bacteria (NOB). The recent discovery of complete ammonia oxidizers (comammox) in the NOB genus Nitrospira, which alone convert ammonia to nitrate, raised questions about the ecological niches in which comammox Nitrospira successfully compete with canonical nitrifiers. Here we isolate a pure culture of a comammox bacterium, Nitrospira inopinata, and show that it is adapted to slow growth in oligotrophic and dynamic habitats on the basis of a high affinity for ammonia, low maximum rate of ammonia oxidation, high growth yield compared to canonical nitrifiers, and genomic potential for alternative metabolisms. The nitrification kinetics of four AOA from soil and hot springs were determined for comparison. Their surprisingly poor substrate affinities and lower growth yields reveal that, in contrast to earlier assumptions, AOA are not necessarily the most competitive ammonia oxidizers present in strongly oligotrophic environments and that N. inopinata has the highest substrate affinity of all analysed ammonia oxidizer isolates except the marine AOA Nitrosopumilus maritimus SCM1 (ref. 3). These results suggest a role for comammox organisms in nitrification under oligotrophic and dynamic conditions.
AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment
2017 - Front Microbiol, 8:1508
Abstract:
Nitrification, the oxidation of ammonia via nitrite to nitrate, has always been considered to be catalyzed by the concerted activity of ammonia- and nitrite-oxidizing microorganisms. Only recently, complete ammonia oxidizers (‘comammox’), which oxidize ammonia to nitrate on their own, were identified in the bacterial genus Nitrospira, previously assumed to contain only canonical nitrite oxidizers. Nitrospira are widespread in nature, but for assessments of the distribution and functional importance of comammox Nitrospira in ecosystems, cultivation-independent tools to distinguish comammox from strictly nitrite oxidizing Nitrospira are required. Here we developed new PCR primer sets that specifically target the amoA genes coding for subunit A of the distinct ammonia monooxygenase of comammox Nitrospira. While existing primers capture only a fraction of the known comammox amoA diversity, the new primer sets cover as much as 95% of the comammox amoA clade A and 92% of the clade B sequences in a reference database containing 326 comammox amoA genes with sequence information at the primer binding sites. Application of the primers to 13 samples from engineered systems (a groundwater well, drinking water treatment and wastewater treatment plants) and other habitats (rice paddy and forest soils, rice rhizosphere, brackish lake sediment and freshwater biofilm) detected comammox Nitrospira in all samples and revealed a considerable diversity of comammox in most habitats. Excellent primer specificity for comammox amoA was achieved by avoiding the use of highly degenerate primer preparations and by using equimolar mixtures of oligonucleotides that match existing comammox amoA genes. Quantitative PCR with these equimolar primer mixtures was highly sensitive and specific, and enabled the efficient quantification of clade A and clade B comammox amoA gene copy numbers in environmental samples. The measured relative abundances of comammox Nitrospira, compared to canonical ammonia oxidizers, were highly variable across environments. The new comammox amoA-targeted primers enable more encompassing future studies of nitrifying microorganisms in diverse habitats. For example, they may be used to monitor the population dynamics of uncultured comammox organisms under changing environmental conditions and in response to altered treatments in engineered and agricultural ecosystems.
Giant viruses with an expanded complement of translation system components.
2017 - Science, 6333: 82-85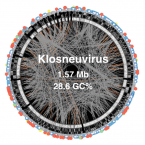
Abstract:
The discovery of giant viruses blurred the sharp division between viruses and cellular life. Giant virus genomes encode proteins considered as signatures of cellular organisms, particularly translation system components, prompting hypotheses that these viruses derived from a fourth domain of cellular life. Here we report the discovery of a group of giant viruses (Klosneuviruses) in metagenomic data. Compared with other giant viruses, the Klosneuviruses encode an expanded translation machinery, including aminoacyl transfer RNA synthetases with specificities for all 20 amino acids. Notwithstanding the prevalence of translation system components, comprehensive phylogenomic analysis of these genes indicates that Klosneuviruses did not evolve from a cellular ancestor but rather are derived from a much smaller virus through extensive gain of host genes.
Crenothrix are major methane consumers in stratified lakes.
2017 - ISME J, 9: 2124-2140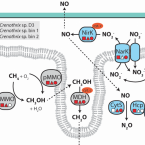
Abstract:
Methane-oxidizing bacteria represent a major biological sink for methane and are thus Earth's natural protection against this potent greenhouse gas. Here we show that in two stratified freshwater lakes a substantial part of upward-diffusing methane was oxidized by filamentous gamma-proteobacteria related to Crenothrix polyspora. These filamentous bacteria have been known as contaminants of drinking water supplies since 1870, but their role in the environmental methane removal has remained unclear. While oxidizing methane, these organisms were assigned an 'unusual' methane monooxygenase (MMO), which was only distantly related to 'classical' MMO of gamma-proteobacterial methanotrophs. We now correct this assignment and show that Crenothrix encode a typical gamma-proteobacterial PmoA. Stable isotope labeling in combination swith single-cell imaging mass spectrometry revealed methane-dependent growth of the lacustrine Crenothrix with oxygen as well as under oxygen-deficient conditions. Crenothrix genomes encoded pathways for the respiration of oxygen as well as for the reduction of nitrate to N2O. The observed abundance and planktonic growth of Crenothrix suggest that these methanotrophs can act as a relevant biological sink for methane in stratified lakes and should be considered in the context of environmental removal of methane.
A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria.
2016 - Trends Microbiol., 9: 699-712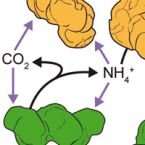
Abstract:
Nitrite-oxidizing bacteria (NOB) catalyze the second step of nitrification, nitrite oxidation to nitrate, which is an important process of the biogeochemical nitrogen cycle. NOB were traditionally perceived as physiologically restricted organisms and were less intensively studied than other nitrogen-cycling microorganisms. This picture is in contrast to new discoveries of an unexpected high diversity of mostly uncultured NOB and a great physiological versatility, which includes complex microbe-microbe interactions and lifestyles outside the nitrogen cycle. Most surprisingly, close relatives to NOB perform complete nitrification (ammonia oxidation to nitrate) and this finding will have far-reaching implications for nitrification research. We review recent work that has changed our perspective on NOB and provides a new basis for future studies on these enigmatic organisms.
Genomics of a phototrophic nitrite oxidizer: insights into the evolution of photosynthesis and nitrification.
2016 - ISME J, 11: 2669-2678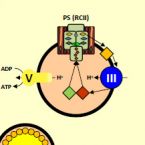
Abstract:
Oxygenic photosynthesis evolved from anoxygenic ancestors before the rise of oxygen ~2.32 billion years ago; however, little is known about this transition. A high redox potential reaction center is a prerequisite for the evolution of the water-oxidizing complex of photosystem II. Therefore, it is likely that high-potential phototrophy originally evolved to oxidize alternative electron donors that utilized simpler redox chemistry, such as nitrite or Mn. To determine whether nitrite could have had a role in the transition to high-potential phototrophy, we sequenced and analyzed the genome of Thiocapsa KS1, a Gammaproteobacteria capable of anoxygenic phototrophic nitrite oxidation. The genome revealed a high metabolic flexibility, which likely allows Thiocapsa KS1 to colonize a great variety of habitats and to persist under fluctuating environmental conditions. We demonstrate that Thiocapsa KS1 does not utilize a high-potential reaction center for phototrophic nitrite oxidation, which suggests that this type of phototrophic nitrite oxidation did not drive the evolution of high-potential phototrophy. In addition, phylogenetic and biochemical analyses of the nitrite oxidoreductase (NXR) from Thiocapsa KS1 illuminate a complex evolutionary history of nitrite oxidation. Our results indicate that the NXR in Thiocapsa originates from a different nitrate reductase clade than the NXRs in chemolithotrophic nitrite oxidizers, suggesting that multiple evolutionary trajectories led to modern nitrite-oxidizing bacteria.
Relative Abundance of Nitrotoga spp. in a Biofilter of a Cold-Freshwater Aquaculture Plant Appears To Be Stimulated by Slightly Acidic pH.
2016 - Appl. Environ. Microbiol., 6: 1838-45
Abstract:
The functioning of recirculation aquaculture systems (RAS) is essential to maintain water quality for fish health, and one crucial process here is nitrification. The investigated RAS was connected to a rainbow trout production system and operated at an average temperature of 13°C and pH 6.8. Community analyses of the nitrifying biofilm revealed a coexistence of Nitrospira and Nitrotoga, and it is hypothesized that a slightly acidic pH in combination with lower temperatures favors the growth of the latter. Modification of the standard cultivation approach toward lower pH values of 5.7 to 6.0 resulted in the successful enrichment (99% purity) of Nitrotoga sp. strain HW29, which had a 16S rRNA sequence similarity of 99.0% to Nitrotoga arctica. Reference cultures of Nitrospira defluvii and the novel Nitrotoga sp. HW29 were used to confirm differentiation of these nitrite oxidizers in distinct ecological niches. Nitrotoga sp. HW29 revealed pH and temperature optima of 6.8 and 22°C, respectively, whereas Nitrospira defluvii displayed the highest nitrite oxidation rate at pH 7.3 and 32°C. We report here the occurrence of Nitrotoga as one of the main nitrite-oxidizing bacteria in freshwater aquaculture systems and indicate that a slightly acidic pH, in addition to temperatures below 20°C, can be applied as a selective isolation criterion for this microorganism.
Complete nitrification by Nitrospira bacteria
2015 - Nature, 528: 504-509
Abstract:
Nitrification, the oxidation of ammonia via nitrite to nitrate, has always been considered to be a two-step process catalysed by chemolithoautotrophic microorganisms oxidizing either ammonia or nitrite. No known nitrifier carries out both steps, although complete nitrification should be energetically advantageous. This functional separation has puzzled microbiologists for a century. Here we report on the discovery and cultivation of a completely nitrifying bacterium from the genus Nitrospira, a globally distributed group of nitrite oxidizers. The genome of this chemolithoautotrophic organism encodes the pathways both for ammonia and nitrite oxidation, which are concomitantly activated during growth by ammonia oxidation to nitrate. Genes affiliated with the phylogenetically distinct ammonia monooxygenase and hydroxylamine dehydrogenase genes of Nitrospira are present in many environments and were retrieved on Nitrospira-contigs in new metagenomes from engineered systems. These findings fundamentally change our picture of nitrification and point to completely nitrifying Nitrospira as key components of nitrogen-cycling microbial communities.
Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira
2015 - Proc Natl Acad Sci U S A, 112: 11371-11376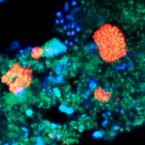
Abstract:
Nitrospira are a diverse group of nitrite-oxidizing bacteria and among the environmentally most widespread nitrifiers. Despite this, they remain scarcely studied and mostly uncultured. Based on genomic and experimental data from Nitrospira moscoviensis representing the ubiquitous Nitrospira lineage II, we identified ecophysiological traits that contribute to the ecological success of Nitrospira. Unexpectedly, N. moscoviensis possesses genes coding for a urease and cleaves urea to ammonia and CO2. Ureolysis was not observed yet in nitrite oxidizers and enables N. moscoviensis to supply ammonia oxidizers lacking urease with ammonia from urea, which is fully nitrified by this consortium through reciprocal feeding. The presence of highly similar urease genes in Nitrospira lenta from activated sludge, in metagenomes from soils and freshwater habitats, and of other ureases in marine nitrite oxidizers, suggests a wide distribution of this extended interaction between ammonia and nitrite oxidizers, which enables nitrite-oxidizing bacteria to indirectly utilize urea as a source of energy. A soluble formate dehydrogenase lends additional ecophysiological flexibility and allows N. moscoviensis to utilize formate, with or without concomitant nitrite oxidation, using oxygen, nitrate, or both compounds as terminal electron acceptors. Compared to Nitrospira defluvii from lineage I, N. moscoviensis shares the Nitrospira core metabolism but shows substantial genomic dissimilarity including genes for adaptations to elevated oxygen concentrations. Reciprocal feeding and metabolic versatility, including the participation in different nitrogen cycling processes, likely are key factors for the niche partitioning, the ubiquity, and the high diversity of Nitrospira in natural and engineered ecosystems.
Abstract:
Ammonia- and nitrite-oxidizing microorganisms are collectively responsible for the aerobic oxidation of ammonia via nitrite to nitrate and have essential roles in the global biogeochemical nitrogen cycle. The physiology of nitrifiers has been intensively studied, and urea and ammonia are the only recognized energy sources that promote the aerobic growth of ammonia-oxidizing bacteria and archaea. Here we report the aerobic growth of a pure culture of the ammonia-oxidizing thaumarchaeote Nitrososphaera gargensis using cyanate as the sole source of energy and reductant; to our knowledge, the first organism known to do so. Cyanate, a potentially important source of reduced nitrogen in aquatic and terrestrial ecosystems, is converted to ammonium and carbon dioxide in Nitrososphaera gargensis by a cyanase enzyme that is induced upon addition of this compound. Within the cyanase gene family, this cyanase is a member of a distinct clade also containing cyanases of nitrite-oxidizing bacteria of the genus Nitrospira. We demonstrate by co-culture experiments that these nitrite oxidizers supply cyanase-lacking ammonia oxidizers with ammonium from cyanate, which is fully nitrified by this microbial consortium through reciprocal feeding. By screening a comprehensive set of more than 3,000 publically available metagenomes from environmental samples, we reveal that cyanase-encoding genes clustering with the cyanases of these nitrifiers are widespread in the environment. Our results demonstrate an unexpected metabolic versatility of nitrifying microorganisms, and suggest a previously unrecognized importance of cyanate in cycling of nitrogen compounds in the environment.
Improved isolation strategies allowed the phenotypic differentiation of two Nitrospira strains from widespread phylogenetic lineages.
2015 - FEMS Microbiol Ecol, 91: fiu031
Abstract:
The second step of nitrification, the oxidation of nitrite to nitrate, is vital for the functioning of the nitrogen cycle, but our understanding of the ecological roles of the involved microorganisms is still limited. The known diversity of Nitrospira, the most widely distributed nitrite-oxidizing bacteria, has increased remarkably by analyses of 16S rRNA and functional gene sequences. However, only few representatives could be brought into laboratory cultures so far. In this study, two Nitrospira from activated sludge were isolated using novel approaches together with established methods. Highly enriched 'Candidatus Nitrospira defluvii' was separated from concomitant heterotrophs by taking advantage of its resistance against ampicillin and acriflavine. Beside this member of lineage I, a novel species of lineage II, named N. lenta, was initially enriched at 10°C and finally purified by using optical tweezers. The tolerance to elevated nitrite levels was much higher in N. defluvii than in the more fastidious N. lenta and was accompanied by pronounced biofilm formation. Phylogenetic classification of 12 additional enrichments indicated that Nitrospira lineage I is common in extreme and moderate ecosystems like lineage II. The new cultures will help to explore physiological and genomic differences affecting niche separation between members of this highly diverse genus.
Dimeric chlorite dismutase from the nitrogen-fixing cyanobacterium Cyanothece sp. PCC7425
2015 - Mol Microbiol, 96: 1053-1068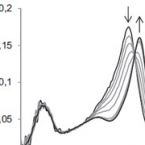
Abstract:
It is demonstrated that cyanobacteria (both azotrophic and non-azotrophic) contain heme b oxidoreductases that can convert chlorite to chloride and molecular oxygen (incorrectly denominated chlorite 'dismutase', Cld). Beside the water-splitting manganese complex of photosystem II, this metalloenzyme is the second known enzyme that catalyses the formation of a covalent oxygen-oxygen bond. All cyanobacterial Clds have a truncated N-terminus and are dimeric (i.e. clade 2) proteins. As model protein, Cld from Cyanothece sp. PCC7425 (CCld) was recombinantly produced in Escherichia coli and shown to efficiently degrade chlorite with an activity optimum at pH 5.0 [kcat 1144 ± 23.8 s-1 , KM 162 ± 10.0 μM, catalytic efficiency (7.1 ± 0.6) × 106 M-1 s-1 ]. The resting ferric high-spin axially symmetric heme enzyme has a standard reduction potential of the Fe(III)/Fe(II) couple of -126 ± 1.9 mV at pH 7.0. Cyanide mediates the formation of a low-spin complex with kon = (1.6 ± 0.1) × 105 M-1 s-1 and koff = 1.4 ± 2.9 s-1 (KD ∼ 8.6 μM). Both, thermal and chemical unfolding follows a non-two-state unfolding pathway with the first transition being related to the release of the prosthetic group. The obtained data are discussed with respect to known structure-function relationships of Clds. We ask for the physiological substrate and putative function of these O2 -producing proteins in (nitrogen-fixing) cyanobacteria.
Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation
2015 - Appl Environ Microbiol, 81: 745-53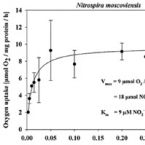
Abstract:
Nitrification has an immense impact on nitrogen cycling in natural ecosystems and in wastewater treatment plants. Mathematical models function as tools to capture the complexity of these biological systems, but kinetic parameters especially of nitrite-oxidizing bacteria (NOB) are lacking because of a limited number of pure cultures until recently. In this study, we compared the nitrite oxidation kinetics of six pure cultures and one enrichment culture representing three genera of NOB (Nitrobacter, Nitrospira, Nitrotoga). With half-saturation constants (Km) between 9 and 27 μM nitrite, Nitrospira bacteria are adapted to live under significant substrate limitation. Nitrobacter showed a wide range of lower substrate affinities, with Km values between 49 and 544 μM nitrite. However, the advantage of Nitrobacter emerged under excess nitrite supply, sustaining high maximum specific activities (Vmax) of 64 to 164 μmol nitrite/mg protein/h, contrary to the lower activities of Nitrospira of 18 to 48 μmol nitrite/mg protein/h. The Vmax (26 μmol nitrite/mg protein/h) and Km (58 μM nitrite) of "Candidatus Nitrotoga arctica" measured at a low temperature of 17°C suggest that Nitrotoga can advantageously compete with other NOB, especially in cold habitats. The kinetic parameters determined represent improved basis values for nitrifying models and will support predictions of community structure and nitrification rates in natural and engineered ecosystems.
Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants
2015 - ISME J, 9: 708-720
Abstract:
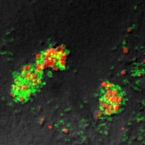
Numerous past studies have shown members of the genus Nitrospira to be the predominant nitrite-oxidizing bacteria (NOB) in nitrifying wastewatertreatment plants (WWTPs). Only recently, the novel NOB 'Candidatus Nitrotoga arctica' was identified in permafrost soil and a close relative was enriched from activated sludge. Still, little is known about diversity, distribution and functional importance of Nitrotoga in natural and engineered ecosystems. Here we developed Nitrotoga 16S rRNA-specific PCR primers and fluorescence in situ hybridization (FISH) probes, which were applied to screen activated sludge samples from 20 full-scale WWTPs. Nitrotoga-like bacteria were detected by PCR in 11 samples and reached abundances detectable by FISH in seven sludges. They coexisted with Nitrospira in most of these WWTPs, but constituted the only detectable NOB in two systems. Quantitative FISH revealed that Nitrotoga accounted for nearly 2% of the total bacterial community in one of these plants, a number comparable to Nitrospira abundances in other WWTPs. Spatial statistics revealed that Nitrotoga coaggregated with ammonia-oxidizing bacteria, strongly supporting a functional role in nitrite oxidation. This activity was confirmed by FISH in combination with microradiography, which revealednitrite-dependent autotrophic carbon fixation by Nitrotoga in situ. Correlation of the presence or absence with WWTP operational parameters indicated low temperatures as a main factor supporting high Nitrotoga abundances, although in incubation experiments these NOB remained active over an unexpected range of temperatures, and also at different ambient nitrite concentrations. In conclusion, this study demonstrates that Nitrotoga can be functionally important nitrite oxidizers in WWTPs and can even represent the only known NOB in engineered systems.
Functionally relevant diversity of closely related Nitrospira in activated sludge
2015 - ISME J, 9: 643-655
Abstract:
Nitrospira are chemolithoautotrophic nitrite-oxidizing bacteria that catalyze the second step of nitrification in most oxic habitats and are important for excess nitrogen removal from sewage in wastewater treatment plants (WWTPs). To date, little is known about their diversity and ecological niche partitioning within complex communities. In this study, the fine-scale community structure and function of Nitrospira was analyzed in two full-scale WWTPs as model ecosystems. In Nitrospira-specific 16S rRNA clone libraries retrieved from each plant, closely related phylogenetic clusters (16S rRNA identities between clusters ranged from 95.8% to 99.6%) within Nitrospira lineages I and II were found. Newly designed probes for fluorescence in situ hybridization (FISH) allowed the specific detection of several of these clusters, whose coexistence in the WWTPs was shown for prolonged periods of several years. In situ ecophysiological analyses based on FISH, relative abundance and spatial arrangement quantification, as well as microautoradiography revealed functional differences of these Nitrospira clusters regarding the preferred nitrite concentration, the utilization of formate as substrate and the spatial coaggregation with ammonia-oxidizing bacteria as symbiotic partners. Amplicon pyrosequencing of the nxrB gene, which encodes subunit beta of nitrite oxidoreductase of Nitrospira, revealed in one of the WWTPs as many as 121 species-level nxrB operational taxonomic units with highly uneven relative abundances in the amplicon library. These results show a previously unrecognized highdiversity of Nitrospira in engineered systems, which is at least partially linked to niche differentiation and may have important implications for process stability.
Structure and heme-binding properties of HemQ (chlorite dismutase-like protein) from Listeria monocytogenes
2015 - Arch Biochem Biophys, 574: 36-48
Abstract:
Chlorite dismutase-like proteins are structurally closely related to functional chlorite dismutases which are heme b-dependent oxidoreductases capable of reducing chlorite to chloride with simultaneous production of dioxygen. Chlorite dismutase-like proteins are incapable of performing this reaction and their biological role is still under discussion. Recently, members of this large protein family were shown to be involved in heme biosynthesis in Gram-positive bacteria, and thus the protein was renamed HemQ in these organisms. In the present work the structural and heme binding properties of the chlorite dismutase-like protein from the Gram-positive pathogen Listeria monocytogenes (LmCld) were analyzed in order to evaluate its potential role as a regulatory heme sensing protein. The homopentameric crystal structure (2.0Å) shows high similarity to chlorite-degrading chlorite dismutases with an important difference in the structure of the putative substrate and heme entrance channel. In solution LmCld is a stable hexamer able to bind the low-spin ligand cyanide. Heme binding is reversible with KD-values determined to be 7.2μM (circular dichroism spectroscopy) and 16.8μM (isothermal titration calorimetry) at pH 7.0. Both acidic and alkaline conditions promote heme release. Presented biochemical and structural data reveal that the chlorite dismutase-like protein from L. monocytogenes could act as a potential regulatory heme sensing and storage protein within heme biosynthesis.
Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium isolated from a bioreactor belonging to the phylum Chloroflexi
2014 - Int J Syst Evol Microbiol, 64: 1859-1865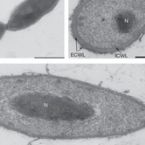
Abstract:
A novel nitrite-oxidizing bacterium (NOB), strain Lb(T), was isolated from a nitrifying bioreactor with a high loading of ammonium bicarbonate in a mineral medium with nitrite as the energy source. The cells were oval (lancet-shaped) rods with pointed edges, non-motile, Gram-positive (by staining and from the cell wall structure) and non-spore-forming. Strain Lb(T) was an obligately aerobic, chemolitoautotrophic NOB, utilizing nitrite or formate as the energy source and CO2 as the carbon source. Ammonium served as the only source of assimilated nitrogen. Growth with nitrite was optimal at pH 6.8-7.5 and at 40 °C (maximum 46 °C). The membrane lipids consisted of C20 alkyl 1,2-diols with the dominant fatty acids being 10MeC18 and C(18 : 1)ω9. The peptidoglycan lacked meso-DAP but contained ornithine and lysine. The dominant lipoquinone was MK-8. Phylogenetic analyses of the 16s rRNA gene sequence placed strain Lb(T) into the class Thermomicrobia of the phylum Chloroflexiwith Sphaerobacter thermophilus as the closest relative. On the basis of physiological and phylogenetic data, it is proposed that strain Lb(T) represents a novel species of a new genus, with the suggested name Nitrolancea hollandica gen. nov., sp. nov. The type strain of the type species is Lb(T) ( = DSM 23161(T) = UNIQEM U798(T)).
Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization.
2014 - Environ Microbiol, 16: 2329-2340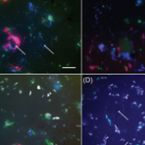
Abstract:
Bacterial colonizers of the aerial parts of plants, or phyllosphere, have been identified on a number of different plants using cultivation-dependent and independent methods. However, the spatial distribution at the micrometer scale of different main phylogenetic lineages is not well documented and mostly based on fluorescence-tagged model strains. In this study, we developed and applied a spatial explicit approach that allowed the use of fluorescence in situ hybridization (FISH) to study bacterial phylloplane communities of environmentally grown Arabidopsis thaliana. We found on average 5.4 × 10(6) bacteria cm(-2) leaf surface and 1.5 × 10(8) bacteria g(-1) fresh weight. Furthermore, we found that the total biomass in the phylloplane was normally distributed. About 31% of the bacteria found in the phylloplane did not hybridize to FISH probes but exhibited infrared autofluorescence indicative for aerobic anoxygenic phototrophs. Four sets of FISH probes targeting Alphaproteobacteria, Betaproteobacteria, Actinobacteria and Bacteroidetes were sufficient to identify all other major contributors of the phylloplane community based on general bacterial probing. Spatial aggregation patterns were observed for all probe-targeted populations at distances up to 7 μm, with stronger tendencies to co-aggregate for members of the same phylogenetic group. Our findings contribute to a bottom-up description of leaf surface community composition.
Thermophilic biological nitrogen removal in industrial wastewater treatment.
2014 - Appl Microbiol Biotechnol, 98: 945-956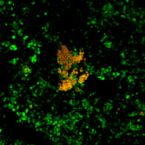
Abstract:
Nitrification is an integral part of biological nitrogen removal processes and usually the limiting step in wastewater treatment systems. Since nitrification is often considered not feasible at temperatures higher than 40 °C, warm industrial effluents (with operating temperatures higher than 40 °C) need to be cooled down prior to biological treatment, which increases the energy and operating costs of the plants for cooling purposes. This study describes the occurrence of thermophilic biological nitrogen removal activity (nitritation, nitratation, and denitrification) at a temperature as high as 50 °C in an activated sludge wastewater treatment plant treating wastewater from an oil refinery. Using a modified two-step nitrification-two-step denitrification mathematical model extended with the incorporation of double Arrhenius equations, the nitrification (nitrititation and nitratation) and denitrification activities were described including the cease in biomass activity at 55 °C. Fluorescence in situ hybridization (FISH) analyses revealed that Nitrosomonas halotolerant and obligatehalophilic and Nitrosomonas oligotropha (known ammonia-oxidizing organisms) and Nitrospira sublineage II (nitrite-oxidizing organism (NOB)) were observed using the FISH probes applied in this study. In particular, this is the first time that Nitrospira sublineage II, a moderatedly thermophilic NOB, is observed in an engineered full-scale (industrial) wastewater treatment system at temperatures as high as 50 °C. These observations suggest that thermophilic biological nitrogen removal can be attained in wastewater treatment systems, which may further contribute to the optimization of the biological nitrogen removal processes in wastewater treatment systems that treat warm wastewaterstreams.
Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation
2014 - Science, 345: 1052-1054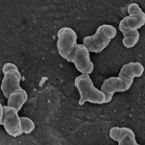
Abstract:
The bacterial oxidation of nitrite to nitrate is a key process of the biogeochemical nitrogen cycle. Nitrite-oxidizing bacteria are considered a highly specialized functional group, which depends on the supply of nitrite from other microorganisms and whose distribution strictly correlates with nitrification in the environment and in wastewater treatment plants. On the basis of genomics, physiological experiments, and single-cell analyses, we show that Nitrospira moscoviensis, which represents a widely distributed lineage of nitrite-oxidizing bacteria, has the genetic inventory to utilizehydrogen (H2) as an alternative energy source for aerobic respiration and grows on H2 without nitrite. CO2 fixation occurred with H2 as the sole electron donor. Our results demonstrate a chemolithoautotrophic lifestyle of nitrite-oxidizing bacteria outside the nitrogen cycle, suggesting greater ecological flexibility than previously assumed.
Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
2014 - Biochemistry, 53: 77-89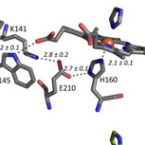
Abstract:
Chlorite dismutases (Clds) are heme b containing oxidoreductases that convert chlorite to chloride and molecular oxygen. In order to elucidate the role of conserved heme cavity residues in the catalysis of this reaction comprehensive mutational and biochemical analyses of Cld from "Candidatus Nitrospira defluvii" (NdCld) were performed. Particularly, point mutations of the cavity-forming residues R173, K141, W145, W146, and E210 were performed. The effect of manipulation in 12 single and double mutants was probed by UV-vis spectroscopy, spectroelectrochemistry, pre-steady-state and steady-state kinetics, and X-ray crystallography. Resulting biochemical data are discussed with respect to the known crystal structure of wild-type NdCld and the variants R173A and R173K as well as the structures of R173E, W145V, W145F, and the R173Q/W146Y solved in this work. The findings allow a critical analysis of the role of these heme cavity residues in the reaction mechanism of chlorite degradation that is proposed to involve hypohalous acid as transient intermediate and formation of an O═O bond. The distal R173 is shown to be important (but not fully essential) for the reaction with chlorite, and, upon addition of cyanide, it acts as a proton acceptor in the formation of the resulting low-spin complex. The proximal H-bonding network including K141-E210-H160 keeps the enzyme in its ferric (E°' = -113 mV) and mainly five-coordinated high-spin state and is very susceptible to perturbation.
Three-dimensional stratification of bacterial biofilm populations in a moving bed biofilm reactor for nitritation anammox.
2014 - Int J Mol Sci, 15: 2191-2206
Abstract:
Moving bed biofilm reactors (MBBRs) are increasingly used for nitrogen removal with nitritation-anaerobic ammonium oxidation (anammox) processes in wastewater treatment. Carriers provide protected surfaces where ammonia oxidizing bacteria (AOB) and anammox bacteria form complex biofilms. However, the knowledge about the organization of microbial communities in MBBR biofilms is sparse. We used new cryosectioning and imaging methods for fluorescence in situ hybridization (FISH) to study the structure of biofilms retrieved from carriers in anitritation-anammox MBBR. The dimensions of the carrier compartments and the biofilm cryosections after FISH showed good correlation, indicating little disturbance of biofilm samples by the treatment. FISH showed that Nitrosomonas europaea/eutropha-related cells dominated the AOB and Candidatus Brocadia fulgida-related cells dominated the anammox guild. New carriers were initially colonized by AOB, followed by anammox bacteria proliferating in the deeper biofilm layers, probably in anaerobic microhabitats created by AOB activity. Mature biofilms showed a pronounced three-dimensional stratification where AOB dominated closer to the biofilm-water interface, whereas anammox were dominant deeper into the carrier space and towards the walls. Our results suggest that current mathematical models may be oversimplifying these three-dimensional systems and unless the multidimensionality of these systems is considered, models may result in suboptimal design of MBBR carriers.
NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira
2014 - Environ Microbiol, 16: 3055-3071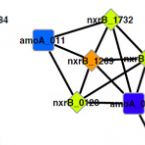
Abstract:
Nitrospira are the most widespread and diverse known nitrite-oxidizing bacteria and key nitrifiers in natural and engineered ecosystems. Nevertheless, their ecophysiology and environmental distribution are understudied due to the recalcitrance of Nitrospira to cultivation and the lack of a molecular functional marker, which would allow the detection of Nitrospira in the environment. Here we introduce nxrB, the gene encoding subunit beta of nitrite oxidoreductase, as a functional and phylogenetic marker for Nitrospira. Phylogenetic trees based on nxrB of Nitrospira were largely congruent to 16S rRNA-based phylogenies. By using new nxrB-selective PCR primers, we obtained almost full-length nxrB sequences from Nitrospira cultures, two activated sludge samples, and several geographically and climatically distinct soils. Amplicon pyrosequencing of nxrB fragments from 16 soils revealed a previously unrecognized diversity of terrestrial Nitrospira with 1,801 detected species-level OTUs (using an inferred species threshold of 95% nxrB identity). Richness estimates ranged from 10 to 946 co-existing Nitrospira species per soil. Comparison to an archaeal amoA dataset obtained from the same soils [Environ. Microbiol. 14: 525-539 (2012)] uncovered that ammonia-oxidizing archaea and Nitrospira communities were highly correlated across the soil samples, possibly indicating shared habitat preferences or specific biological interactions among members of these nitrifier groups.
Enrichment and genome sequence of the group I.1a ammonia-oxidizing archaeon "Ca. Nitrosotenuis uzonensis" representing a clade globally distributed in thermal habitats
2013 - PLoS One, 8: e80835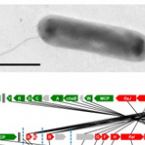
Abstract:
The discovery of ammonia-oxidizing archaea (AOA) of the phylum Thaumarchaeota and the high abundance of archaeal ammonia monooxygenase subunit A encoding gene sequences in many environments have extended our perception of nitrifying microbial communities. Moreover, AOA are the only aerobic ammonia oxidizers known to be active in geothermal environments. Molecular data indicate that in many globally distributed terrestrial high-temperature habits a thaumarchaeotal lineage within the Nitrosopumilus cluster (also called marine group I.1a) thrives, but these microbes have neither been isolated from these systems nor functionally characterized in situ yet. In this study, we report on the enrichment and genomic characterization of a representative of this lineage from a thermal spring in Kamchatka. This thaumarchaeote, provisionally classified as "Candidatus Nitrosotenuis uzonensis", is a moderately thermophilic, non-halophilic, chemolithoautotrophic ammonia oxidizer. The nearly complete genome sequence (assembled into a single scaffold) of this AOA confirmed the presence of the typical thaumarchaeotal pathways for ammonia oxidation and carbon fixation, and indicated its ability to produce coenzyme F420 and to chemotactically react to its environment. Interestingly, like members of the genus Nitrosoarchaeum, "Candidatus N. uzonensis" also possesses a putative artubulin-encoding gene. Genome comparisons to related AOA with available genome sequences confirmed that the newly cultured AOA has an average nucleotide identity far below the species threshold and revealed a substantial degree of genomic plasticity with unique genomic regions in "Ca. N. uzonensis", which potentially include genetic determinants of ecological niche differentiation.
New methods for analysis of spatial distribution and co-aggregation of microbial populations in complex biofilms
2013 - Appl Environ Microbiol, 79: 5978-5987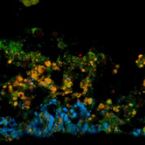
Abstract:
In biofilms, microbial activities form gradients of substrates and electron acceptors, creating a complex landscape of microhabitats, often resulting in structured localization of the microbial populations present. To understand the dynamic interplay between and within these populations, quantitative measurements and statistical analysis of their localization patterns within the biofilms are necessary, and adequate automated tools for such analyses are needed. We have designed and applied new methods for fluorescence in situ hybridization (FISH) and digital image analysis of directionally dependent (anisotropic) multispecies biofilms. A sequential-FISH approach allowed multiple populations to be detected in a biofilm sample. This was combined with an automated tool for vertical-distribution analysis by generating in silico biofilm slices and the recently developed Inflate algorithm for coaggregation analysis of microbial populations in anisotropic biofilms. As a proof of principle, we show distinct stratification patterns of the ammonia oxidizers Nitrosomonas oligotropha subclusters I and II and the nitrite oxidizer Nitrospira sublineage I in three different types of wastewater biofilms, suggesting niche differentiation between the N. oligotropha subclusters, which could explain their coexistence in the same biofilms. Coaggregation analysis showed that N. oligotropha subcluster II aggregated closer to Nitrospira than did N. oligotropha subcluster I in a pilot plant nitrifying trickling filter (NTF) and a moving-bed biofilm reactor (MBBR), but not in a full-scale NTF, indicating important ecophysiological differences between these phylogenetically closely related subclusters. By using high-resolution quantitative methods applicable to any multispecies biofilm in general, the ecological interactions of these complex ecosystems can be understood in more detail.
Colonization of freshwater biofilms by nitrifying bacteria from activated sludge
2013 - FEMS Microbiol Ecol, 85: 104-115
Abstract:
Effluents from wastewater treatment plants (WWTPs) containing micro-organisms and residual nitrogen can stimulate nitrification in freshwater streams. We hypothesized that different ammonia-oxidizing (AOB) and nitrite-oxidizing (NOB) bacteria present in WWTP effluents differ in their potential to colonize biofilms in the receiving streams. In an experimental approach, we monitored biofilm colonization by nitrifiers in ammonium- or nitrite-fed microcosm flumes after inoculation with activated sludge. In a field study, we compared the nitrifier communities in a full-scale WWTP and in epilithic biofilms downstream of the WWTP outlet. Despite substantially different ammonia concentrations in the microcosms and the stream, the same nitrifiers were detected by fluorescence in situ hybridization in all biofilms. Of the diverse nitrifiers present in the WWTPs, only AOB of the Nitrosomonas oligotropha/ureae lineage and NOB of Nitrospira sublineage I colonized the natural biofilms. Analysis of the amoA gene encoding the alpha subunit of ammonia monooxygenase of AOB revealed seven identical amoA sequence types. Six of these affiliated with the N. oligotropha/ureae lineage and were shared between the WWTP and the stream biofilms, but the other shared sequence type grouped with the N. europaea/eutropha and N. communis lineage. Measured nitrification activities were high in the microcosms and the stream. Our results show that nitrifiers from WWTPs can colonize freshwater biofilms and confirm that WWTP-affected streams are hot spots of nitrification.
The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer
2013 - Front Microbiol, 4: 27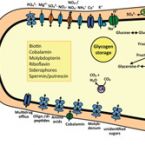
Abstract:
In marine systems, nitrate is the major reservoir of inorganic fixed nitrogen. The only known biological nitrate-forming reaction is nitrite oxidation, but despite its importance, our knowledge of the organisms catalyzing this key process in the marine N-cycle is very limited. The most frequently encountered marine NOB are related to Nitrospina gracilis, an aerobic chemolithoautotrophic bacterium isolated from ocean surface waters. To date, limited physiological and genomic data for this organism were available and its phylogenetic affiliation was uncertain. In this study, the draft genome sequence of N. gracilis strain 3/211 was obtained. Unexpectedly for an aerobic organism, N. gracilis lacks classical reactive oxygen defense mechanisms and uses the reductive tricarboxylic acid cycle for carbon fixation. These features indicate microaerophilic ancestry and are consistent with the presence of Nitrospina in marine oxygen minimum zones. Fixed carbon is stored intracellularly as glycogen, but genes for utilizing external organic carbon sources were not identified. N. gracilis also contains a full gene set for oxidative phosphorylation with oxygen as terminal electron acceptor and for reverse electron transport from nitrite to NADH. A novel variation of complex I may catalyze the required reverse electron flow to low-potential ferredoxin. Interestingly, comparative genomics indicated a strong evolutionary link between Nitrospina, the nitrite-oxidizing genus Nitrospira, and anaerobic ammonium oxidizers, apparently including the horizontal transfer of a periplasmically oriented nitrite oxidoreductase and other key genes for nitrite oxidation at an early evolutionary stage. Further, detailed phylogenetic analyses using concatenated marker genes provided evidence that Nitrospina forms a novel bacterial phylum, for which we propose the name Nitrospinae.
Interactions of nitrifying bacteria and heterotrophs: Identification of a Micavibrio-like, putative predator of Nitrospira
2013 - Appl Environ Microbiol, 79: 2027-2037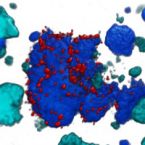
Abstract:
Chemolithoautotrophic nitrifying bacteria release soluble organic compounds, which can be substrates for heterotrophic microorganisms. The identity of these heterotrophs and the specificity of their interactions with nitrifiers are largely unknown. Here we incubated nitrifying activated sludge with (13)C-labeled bicarbonate and used stable isotope probing of 16S rRNA to follow the flow of carbon from uncultured nitrifiers to heterotrophs. To facilitate the identification of heterotrophs, the abundant 16S rRNA molecules from nitrifiers were depleted by catalytic oligonucleotides containing locked nucleic acids (LNAzymes), which specifically cut the 16S rRNA of defined target organisms. Among the (13)C-labeled heterotrophs were organisms remotely related to Micavibrio, a microbial predator of Gram-negative bacteria. Fluorescence in situ hybridization revealed a close spatial association of these organisms with microcolonies of nitrite-oxidizing sublineage I Nitrospira in sludge flocs. The high specificity of this interaction was confirmed by confocal microscopy and a novel image analysis method to quantify the localization patterns of biofilm microorganisms in 3-D space. Other isotope-labeled bacteria, which were affiliated with Thermomonas, co-localized less frequently with nitrifiers and thus were commensals or saprophytes rather than specific symbionts or predators. These results suggest that Nitrospira are subject to bacterial predation, which may influence the abundance and diversity of these nitrite oxidizers and the stability of nitrification in engineered and natural ecosystems. In silico screening of published next-generation sequencing datasets revealed a broad environmental distribution of the uncultured Micavibrio-like lineage.
Structure and community composition of sprout-like bacterial aggregates in a Dinaric karst subterraenean stream
2013 - Microb. Ecol., 66: 5-18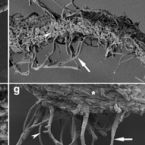
Abstract:
The Vjetrenica cave in the Dinaric Karst hosts a worldwide extraordinarily high cave biodiversity. Beside a diverse and specialized cave fauna, sprout-like formations attached to the bed of the cave stream were observed and described, but not further characterized, almost a century ago. Here we investigated these sprout-like microbial aggregates by the rRNA approach and detailed microscopy. Based on fluorescence in situ hybridization and ultrastructural analysis, the sprout-like formations are morphologically highly organized and their core consists of a member of a novel deep-branching lineage in the bacterial phylum Nitrospirae. This organism displays an interesting cellular ultrastructure with different kinds of cytoplasmic inclusions and is embedded in a thick extracellular matrix, which contributes to the stability and shape of the aggregates. This novel bacterium has been provisionally classified as ‘Candidatus Troglogloea absoloni’. The surface of the sprout-like aggregates is more diverse than the core. It is colonized by a bacterial biofilm consisting primarily of filamentous Betaproteobacteria, whereas other microbial populations present in the crust include members of the Bacteriodetes, Gammaproteobacteria, Actinombacteria, Alphaproteobacteria and Planctomycetes, which are intermingled with mineral inclusions. This study represents the first thorough molecular and ultrastructural characterization of the elusive sprout-like bacterial aggregates, which are also found in other cave systems of the Dinaric karst. The discovery of Ca. Troglogloea absoloni contributes to the known biodiversity of subterranean ecosystems and especially of macroscopic structures formed in caves by microorganisms, whose composition and ecological function often remain enigmatic.
Depletion of unwanted nucleic acid templates by selective cleavage: LNAzymes open a new window for detecting rare microbial community members
2013 - Appl. Environ. Microbiol., 79: 1534-1544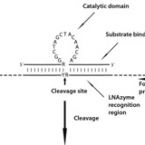
Abstract:
Many studies of molecular microbial ecology rely on the characterization of microbial communities by PCR amplification, cloning, sequencing, and phylogenetic analysis of genes encoding rRNAs or functional marker enzymes. However, if the established clone libraries are dominated by one or a few sequence types, the cloned diversity is difficult to analyze by random clone sequencing. Here we present a novel approach to deplete unwanted sequence types from complex nucleic acid mixtures prior to cloning and downstream analyses. It employs catalytically active oligonucleotides containing locked nucleic acids (LNAzymes) for the specific cleavage of selected RNA targets. When combined with in vitro transcription and reverse transcriptase PCR, this LNAzyme-based technique can be used with DNA or RNA extracts from microbial communities. The simultaneous application of more than one specific LNAzyme allows the concurrent depletion of different sequence types from the same nucleic acid preparation. This new method was evaluated with defined mixtures of cloned 16S rRNA genes and then used to identify accompanying bacteria in an enrichment culture dominated by the nitrite oxidizer "Candidatus Nitrospira defluvii." In silico analysis revealed that the majority of publicly deposited rRNA-targeted oligonucleotide probes may be used as specific LNAzymes with no or only minor sequence modifications. This efficient and cost-effective approach will greatly facilitate tasks such as the identification of microbial symbionts in nucleic acid preparations dominated by plastid or mitochondrial rRNA genes from eukaryotic hosts, the detection of contaminants in microbial cultures, and the analysis of rare organisms in microbial communities of highly uneven composition.
Nitrogen processing and the role of stream benthic biofilms downstream of a wastewater treatment plant
2012 - Freshwater Science, 31: 1057-1069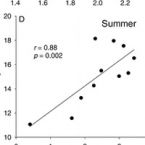
Abstract:
We investigated how dissolved inorganic N (DIN) inputs from a wastewater treatment plant (WWTP) effluent are processed biogeochemically by the receiving stream. We examined longitudinal patterns of NH4+ and NO3− concentrations and their 15N signatures along a stream reach downstream of a WWTP. We compared the δ15N signatures of epilithic biofilms with those of DIN to assess the role of stream biofilms in N processing. We analyzed the δ15N signatures of biofilms coating light- and dark-side surfaces of cobbles separately to test whether light constrains functioning of biofilm communities. We sampled during 2 contrasting periods of the year (winter and summer) to explore whether changes in environmental conditions affected N biogeochemical processes. The study reach had a remarkable capacity for transformation and removal of DIN, but the magnitude and relevance of different biogeochemical pathways of N processing differed between seasons. In winter, assimilation and nitrification influenced downstream N fluxes. These processes were spatially segregated at the microhabitat scale, as indicated by a significant difference in the δ15N signature of light- and dark-side biofilms, a result suggesting that nitrification was mostly associated with dark-side biofilms. In summer, N processing was intensified, and denitrification became an important N removal pathway. The δ15N signatures of the light- and dark-side biofilms were similar, a result suggesting less spatial segregation of N cycling processes at this microhabitat scale. Collectively, our results highlight the capacity of WWTP-influenced streams to transform and remove WWTP-derived N inputs and indicate the active role of biofilms in these in-stream processes.
Redox thermodynamics of high-spin and low-spin forms of chlorite dismutases of diverse subunit and oligomeric structure
2012 - Biochemistry, 51: 9501-9512
Abstract:
Chlorite dismutases (Clds) are heme b-containing oxidoreductases that convert chlorite to chloride and dioxygen. In this work, the thermodynamics of the one-electron reduction of the ferric high-spin forms and of the six-coordinate low-spin cyanide adducts of the enzymes from Nitrobacter winogradskyi (NwCld) and Candidatus "Nitrospira defluvii" (NdCld) were determined through spectroelectrochemical experiments. These proteins belong to two phylogenetically separated lineages that differ in subunit (21.5 and 26 kDa, respectively) and oligomeric (dimeric and pentameric, respectively) structure but exhibit similar chlorite degradation activity. The E°' values for free and cyanide-bound proteins were determined to be -119 and -397 mV for NwCld and -113 and -404 mV for NdCld, respectively (pH 7.0, 25 °C). Variable-temperature spectroelectrochemical experiments revealed that the oxidized state of both proteins is enthalpically stabilized. Molecular dynamics simulations suggest that changes in the protein structure are negligible, whereas solvent reorganization is mainly responsible for the increase in entropy during the redox reaction. Obtained data are discussed with respect to the known structures of the two Clds and the proposed reaction mechanism.
Nitrification expanded: Discovery, physiology, and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi
2012 - ISME J., 6: 2245-2256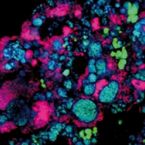
Abstract:
Nitrite-oxidizing bacteria (NOB) catalyze the second step of nitrification, a major process of the biogeochemical nitrogen cycle, but the recognized diversity of this guild is surprisingly low and only two bacterial phyla contain known NOB. Here, we report on the discovery of a chemolithoautotrophic nitrite oxidizer that belongs to the widespread phylum Chloroflexi not previously known to contain any nitrifying organism. This organism, named Nitrolancetus hollandicus, was isolated from a nitrifying reactor. Its tolerance to a broad temperature range (25-63 °C) and low affinity for nitrite (K(s)=1 mM), a complex layered cell envelope that stains Gram positive, and uncommon membrane lipids composed of 1,2-diols distinguish N. hollandicus from all other known nitrite oxidizers. N. hollandicus grows on nitrite and CO(2), and is able to use formate as a source of energy and carbon. Genome sequencing and analysis of N. hollandicus revealed the presence of all genes required for CO(2) fixation by the Calvin cycle and a nitrite oxidoreductase (NXR) similar to the NXR forms of the proteobacterial nitrite oxidizers, Nitrobacter and Nitrococcus. Comparative genomic analysis of the nxr loci unexpectedly indicated functionally important lateral gene transfer events between Nitrolancetus and other NOB carrying a cytoplasmic NXR, suggesting that horizontal transfer of the NXR module was a major driver for the spread of the capability to gain energy from nitrite oxidation during bacterial evolution. The surprising discovery of N. hollandicus significantly extends the known diversity of nitrifying organisms and likely will have implications for future research on nitrification in natural and engineered ecosystems.
Impact of subunit and oligomeric structure on the thermal and conformational stability of chlorite dismutases
2012 - Biochim. Biophys. Acta, 1824: 1031-1038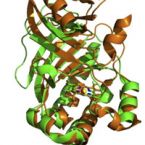
Abstract:
Chlorite dismutases (Cld) are unique heme b containing oxidoreductases that convert chlorite to chloride and dioxygen. Recent phylogenetic and structural analyses demonstrated that these metalloproteins significantly differ in oligomeric and subunit structure. Here we have analyzed two representatives of two phylogenetically separated lineages, namely pentameric Cld from Candidatus "Nitrospira defluvii" and dimeric Cld from Nitrobacter winogradskyi having a similar enzymatic activity at room temperature. By application of a broad set of techniques including differential scanning calorimetry, electronic circular dichroism, UV-vis and fluorescence spectroscopy the temperature-mediated and chemical unfolding of both recombinant proteins were analyzed. Significant differences in thermal and conformational stability are reported. The pentameric enzyme is very stable between pH 3 and 10 (T(m)=92°C at pH 7.0) and active at high temperatures thus being an interesting candidate for bioremediation of chlorite. By contrast the dimeric protein starts to unfold already at 53°C. The observed unfolding pathways are discussed with respect to the known subunit structure and subunit interaction.
Co-localized or randomly distributed? Pair cross correlation of in vivo grown subgingival biofilm bacteria quantified by digital image analysis
2012 - PLoS One, 7: e37583
Abstract:
The polymicrobial nature of periodontal diseases is reflected by the diversity of phylotypes detected in subgingival plaque and the finding that consortia of suspected pathogens rather than single species are associated with disease development. A number of these microorganisms have been demonstrated in vitro to interact and enhance biofilm integration, survival or even pathogenic features. To examine the in vivo relevance of these proposed interactions, we extended the spatial arrangement analysis tool of the software daime (digital image analysis in microbial ecology). This modification enabled the quantitative analysis of microbial co-localization in images of subgingival biofilm species, where the biomass was confined to fractions of the whole-image area, a situation common for medical samples. Selected representatives of the disease-associated red and orange complexes that were previously suggested to interact with each other in vitro (Tannerella forsythia with Fusobacterium nucleatum and Porphyromonas gingivalis with Prevotella intermedia) were chosen for analysis and labeled with specific fluorescent probes via fluorescence in situ hybridization. Pair cross-correlation analysis of in vivo grown biofilms revealed tight clustering of F. nucleatum/periodonticum and T. forsythia at short distances (up to 6 µm) with a pronounced peak at 1.5 µm. While these results confirmed previous in vitro observations for F. nucleatum and T. forsythia, random spatial distribution was detected between P. gingivalis and P. intermedia in the in vivo samples. In conclusion, we successfully employed spatial arrangement analysis on the single cell level in clinically relevant medical samples and demonstrated the utility of this approach for the in vivo validation of in vitro observations by analyzing statistically relevant numbers of different patients. More importantly, the culture-independent nature of this approach enables similar quantitative analyses for as-yet-uncultured phylotypes which cannot be characterized in vitro.
Linking microbial and ecosystem ecology using ecological stoichiometry: A synthesis of conceptual and empirical approaches
2011 - Ecosystems, 14: 261-273
Abstract:
Currently, one of the biggest challenges in microbial and ecosystem ecology is to develop conceptual models that organize the growing body of information on environmental microbiology into a clear mechanistic framework with a direct link to ecosystem processes. Doing so will enable development of testable hypotheses to better direct future research and increase understanding of key constraints on biogeochemical networks. Although the understanding of phenotypic and genotypic diversity of microorganisms in the environment is rapidly accumulating, how controls on microbial physiology ultimately affect biogeochemical fluxes remains poorly understood. We propose that insight into constraints on biogeochemical cycles can be achieved by a more rigorous evaluation of microbial community biomass composition within the context of ecological stoichiometry. Multiple recent studies have pointed to microbial biomass stoichiometry as an important determinant of when microorganisms retain or recycle mineral nutrients. We identify the relevant cellular components that most likely drive changes in microbial biomass stoichiometry by defining a conceptual model rooted in ecological stoichiometry. More importantly, we show how X-ray microanalysis (XRMA), nanoscale secondary ion mass spectroscopy (NanoSIMS), Raman microspectroscopy, and in situ hybridization techniques (for example, FISH) can be applied in concert to allow for direct empirical evaluation of the proposed conceptual framework. This approach links an important piece of the ecological literature, ecological stoichiometry, with the molecular front of the microbial revolution, in an attempt to provide new insight into how microbial physiology could constrain ecosystem processes.
Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers
2011 - Proc. Natl. Acad. Sci. USA, 108: 16771-16776
Abstract:
Nitrification is a core process in the global nitrogen cycle that is essential for the functioning of many ecosystems. The discovery of autotrophic ammonia-oxidizing archaea (AOA) within the phylum Thaumarchaeota has changed our perception of the microbiology of nitrification, in particular since their numerical dominance over ammonia-oxidizing bacteria (AOB) in many environments has been revealed. These and other data have led to a widely held assumption that all amoA-encoding members of the Thaumarchaeota (AEA) are autotrophic nitrifiers. In this study, 52 municipal and industrial wastewater treatment plants were screened for the presence of AEA and AOB. Thaumarchaeota carrying amoA were detected in high abundance only in four industrial plants. In one plant, thaumarchaeotes closely related to soil group I.1b outnumbered AOB up to 10,000-fold, and their numbers, which can only be explained by active growth in this continuous culture system, were two to three orders of magnitude higher than could be sustained by autotrophic ammonia oxidation. Consistently, (14)CO(2) fixation could only be detected in AOB but not in AEA in actively nitrifying sludge from this plant via FISH combined with microautoradiography. Furthermore, in situ transcription of archaeal amoA, and very weak in situ labeling of crenarchaeol after addition of (13)CO(2), was independent of the addition of ammonium. These data demonstrate that some amoA-carrying group I.1b Thaumarchaeota are not obligate chemolithoautotrophs.
In situ techniques and digital image analysis methods for quantifying spatial localization patterns of nitrifiers and other microorganisms in biofilm and flocs
2011 - Methods Enzymol., 496: 185-215
Abstract:
The spatial localization patterns of microorganisms in multispecies biofilms reflect numerous phenomena that influence sessile microbial life, such as substrate concentration gradients within the biofilm and biological interactions with other biofilm populations. Quantitative and population-specific in situ analyses of spatial patterns have a high potential to provide novel insights into the biology of biofilm organisms, including yet uncultured microbes, but such approaches have been developed and used in a few studies only. Here, we outline digital image analysis methods to quantify the coaggregation, mutual avoidance, or random distribution of microbial populations in biofilm and flocs. A protocol is provided for fluorescence in situ hybridization with rRNA-targeted probes, which preserves the three-dimensional biofilm architecture for confocal microscopy and image analysis, and the combined use of these approaches is demonstrated by spatial analyses of nitrifying bacteria in complex biofilm samples.
Unexpected diversity of chlorite dismutases: A catalytically efficient dimeric enzyme from Nitrobacter winogradskyi
2011 - J. Bacteriol., 193: 2408-2417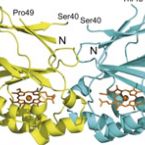
Abstract:
Chlorite dismutase (Cld) is a unique heme enzyme catalyzing the conversion of ClO2- to Cl- and O2. Cld is usually found in perchlorate- or chlorate-reducing bacteria, but was recently identified also in a nitrite-oxidizing bacterium of the genus Nitrospira. Here we characterized a novel Cld-like protein from the chemolithoautotrophic nitrite oxidizer Nitrobacter winogradskyi, which is significantly smaller than all previously known chlorite dismutases. Its 3D crystal structure revealed a dimer of two identical subunits, which sharply contrasts the penta- or hexameric structures of other chlorite dismutases. Despite a truncated N-terminal domain in each subunit, this novel enzyme turned out to be a highly efficient chlorite dismutase (KM = 90 µM, kcat = 190 s-1, kcat/KM = 2.1 106 M-1 s-1), demonstrating a greater structural and phylogenetic diversity of these enzymes than previously known. Based on comparative analyses of Cld sequences and 3D structures, signature amino acid residues were identified that can be employed to assess whether uncharacterized Cld-like proteins may have a high chlorite-dismutating activity. Interestingly, proteins that contain all these signatures and are phylogenetically closely related to the novel-type Cld of N. winogradskyi exist in a large number of other microbes, including other nitrite oxidizers.
Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring
2011 - FEMS Microbiol. Ecol., 75: 195-204
Abstract:
Geothermal environments are a suitable habitat for nitrifying microorganisms. Conventional and molecular techniques indicated that chemolithoautotrophic nitrite-oxidizing bacteria affiliated with the genus Nitrospira are widespread in environments with elevated temperatures up to 55 °C in Asia, Europe, and Australia. However, until now, no thermophilic pure cultures of Nitrospira were available, and the physiology of these bacteria was mostly uncharacterized. Here, we report on the isolation and characterization of a novel thermophilic Nitrospira strain from a microbial mat of the terrestrial geothermal spring Gorjachinsk (pH 8.6; temperature 48 °C) from the Baikal rift zone (Russia). Based on phenotypic properties, chemotaxonomic data, and 16S rRNA gene phylogeny, the isolate was assigned to the genus Nitrospira as a representative of a novel species, for which the name Nitrospira calida is proposed. A highly similar 16S rRNA gene sequence (99.6% similarity) was detected in a Garga spring enrichment grown at 46 °C, whereas three further thermophilic Nitrospira enrichments from the Garga spring and from a Kamchatka Peninsula (Russia) terrestrial hot spring could be clearly distinguished from N. calida (93.6-96.1% 16S rRNA gene sequence similarity). The findings confirmed that Nitrospira drive nitrite oxidation in moderate thermophilic habitats and also indicated an unexpected diversity of heat-adapted Nitrospira in geothermal hot springs.
Looking inside the box: Using Raman microspectroscopy to deconstruct microbial biomass stoichiometry one cell at a time
2011 - ISME J., 5: 196-208
Abstract:
Stoichiometry of microbial biomass is a key determinant of nutrient recycling in a wide variety of ecosystems. However, little is known about the underlying causes of variance in microbial biomass stoichiometry. This is primarily because of technological constraints limiting the analysis of macromolecular composition to large quantities of microbial biomass. Here, we use Raman microspectroscopy (MS), to analyze the macromolecular composition of single cells of two species of bacteria grown on minimal media over a wide range of resource stoichiometry. We show that macromolecular composition, determined from a subset of identified peaks within the Raman spectra, was consistent with macromolecular composition determined using traditional analytical methods. In addition, macromolecular composition determined by Raman MS correlated with total biomass stoichiometry, indicating that analysis with Raman MS included a large proportion of a cell's total macromolecular composition. Growth phase (logarithmic or stationary), resource stoichiometry and species identity each influenced each organism's macromolecular composition and thus biomass stoichiometry. Interestingly, the least variable peaks in the Raman spectra were those responsible for differentiation between species, suggesting a phylogenetically specific cellular architecture. As Raman MS has been previously shown to be applicable to cells sampled directly from complex environments, our results suggest Raman MS is an extremely useful application for evaluating the biomass stoichiometry of environmental microorganisms. This includes the ability to partition microbial biomass into its constituent macromolecules and increase our understanding of how microorganisms in the environment respond to resource heterogeneity.
A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria
2010 - Proc. Natl. Acad. Sci. USA, 107: 13479-13484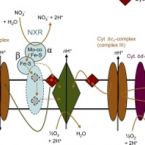
Abstract:
Nitrospira are barely studied and mostly uncultured nitrite-oxidizing bacteria, which are according to molecular data among the most diverse and widespread nitrifiers in natural ecosystems and biological wastewater treatment. Here, environmental genomics was used to reconstruct the complete genome of "Candidatus Nitrospira defluvii" from an activated sludge enrichment culture. Based on this first deciphered Nitrospira genome and on experimental data, we show that Ca. N. defluvii differs dramatically from other known nitrite oxidizers in the key enzyme nitrite oxidoreductase (NXR), the composition of the respiratory chain, and the pathway used for autotrophic carbon fixation, suggesting multiple independent evolution of chemolithoautotrophic nitrite oxidation. Adaptations of Ca. N. defluvii to substrate-limited conditions include a novel periplasmic NXR, which is constitutively expressed, and pathways for the transport, oxidation and assimilation of simple organic compounds that allow a mixotrophic lifestyle. The reverse tricarboxylic acid cycle as pathway for CO2 fixation and the lack of most classical defence mechanisms against oxidative stress suggest that Nitrospira evolved from microaerophilic or even anaerobic ancestors. Unexpectedly, comparative genomic analyses indicate functionally significant lateral gene transfer events between the genus Nitrospira and anaerobic ammonium-oxidizing planctomycetes, which share highly similar forms of NXR and other proteins reflecting that two key processes of the nitrogen cycle are evolutionary connected.
Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
2010 - J. Struct. Biol., 172: 331-342
Abstract:
Chlorite dismutase (Cld) is a unique heme enzyme which transforms chlorite to chloride and molecular oxygen (reaction: ClO(2)(-)→Cl(-)+O(2)). Since bacteria with Cld play significant roles in the bioremediation of industrially contaminated sites and also in wastewater treatment, it is of high interest to understand the molecular mechanism of chlorite detoxification. Here we investigate a highly active Cld from Candidatus Nitrospira defluvii (NdCld), a key nitrifier in biological wastewater treatment, using a comprehensive structural, biochemical and bioinformatics approach. We determined the crystal structure of Cld from Candidatus Nitrospira defluvii and showed that functional NdCld is a homopentamer possessing a fold found in other Clds and Cld-like enzymes. To investigate the Cld function in more detail, site-directed mutagenesis of a catalytically important residue (Arg173) was performed and two enzyme mutants were structurally and biochemically characterized. Arginine 173 is demonstrated to play a key role in (i) controlling of ligand and substrate access and binding and (ii) in chlorite dismutation reaction. The flexible residue modulates the electrostatic potential and size of the active site entrance and might be involved in keeping transiently formed hypochlorite in place for final molecular oxygen and chloride formation. Furthermore, using a structure-based sequence alignment, we show that the residue corresponding to Arg173 is conserved in all known active forms of Cld and propose it as a marker for Cld activity in yet uncharacterized Cld-like proteins. Finally, our analysis indicates that all Clds and Cld-like enzymes employ a non-covalently bound heme as a cofactor.
Drivers of bacterial colonization patterns in stream biofilms
2010 - FEMS Microbiol. Ecol., 72: 47-57
Abstract:
Dispersal and colonization are important for the assembly and biodiversity of microbial communities. While emigration as the initial step of dispersal has become increasingly understood in model bacterial biofilms, the drivers of dispersal and colonization in complex biofilms remain elusive. We grew complex biofilms in microcosms from natural surface water in laminar and turbulent flow, and investigated dispersal and colonization patterns of fluorescently labeled cells and microbeads in nascent and mature biofilms. Settling occurred in nonrandom spatial patterns governed by the interplay of local flow patterns and biofilm topography. Settling was higher in treatments with nascent biofilms, with fewer cells remaining in the water column than in treatments with mature biofilms. The flow regime had no effect on settling velocity, even though in mature biofilms the formation of streamers under turbulent flow enhanced particle trapping compared with the laminar flow treatment. Hence, small-scale variations in the flow pattern seemed to be more important than the overall flow regime. Furthermore, spatial analysis of the colonizer patterns suggests that bacteria have moved in the biofilm after settling. Our results show that colonization of biofilms in a model stream environment is a heterogeneous process differently affected by biological and physical factors.
Double-labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility
2010 - Appl. Environ. Microbiol., 76: 922–926
Abstract:
Fluorescence in situ hybridization (FISH) with singly labeled rRNA-targeted oligonucleotide probes is widely applied for direct identification of microbes in the environment or in clinical specimens. Here we show that a replacement of singly labeled oligonucleotide probes with 5'-, 3'-doubly labeled probes at least doubles FISH signal intensity without causing specificity problems. Furthermore, Cy3-doubly labeled probes strongly increase in situ accessibility of rRNA target sites and thus provide more flexibility for probe design.
Use of fluorescence in situ hybridization and the daime image analysis program for the cultivation-independent quantification of microorganisms in environmental and medical samples
2009 - Cold Spring Harb. Protoc., 4: 10.1101/pdb.prot5253
Abstract:
Conventional cultivation-based methods to measure microbial abundance are unsuitable for quantifying uncultured microorganisms that constitute the majority of microbial life in most environmental or medical samples. This problem is solved by the quantification approach described here, which combines fluorescence in situ hybridization (FISH) with rRNA-targeted probes and digital image analysis. By measuring the areas of probe-labeled biomass in randomly recorded image pairs, an unbiased estimate of the relative biovolume of the population of interest can be obtained. This approach expresses abundance as "biovolume fraction" (relative to the total biovolume of the whole microbial community). This value equals the share of biochemical reaction space occupied by the quantified population and thus can be more relevant ecologically than absolute cell numbers (e.g., a few large cells can contain the same biovolume as many small cells). Another advantage lies in the complete independence of this method from the morphology of the quantified organisms. Regardless of whether the target microbes occur as single cells in plankton samples, as filaments, or as dense aggregates in biofilms, this cultivation-independent method allows the composition of complex microbial communities to be determined
Initial effects of experimental warming on carbon exchange rates, plant growth and microbial dynamics of a lichen-rich dwarf shrub tundra in Siberia
2008 - Plant Soil, 307: 191-205
Abstract:
The aim of this study was to assess initial effects of warming on the CO2 balance of a lichen-rich dwarf shrub tundra, a widespread but little studied ecosystem type in the Arctic. We analyzed whole ecosystem carbon exchange rates as well as nutrient dynamics, microbial and plant community composition and biomass after 2 years of experimental temperature increase. Plant biomass increased significantly with warming, mainly due to the strong response of lichens, the dominant plant group within this ecosystem. Experimental warming also increased soil nitrogen pools and nitrogen turnover rates. Major changes in soil microbial and plant composition, however, were not detected. Although experimental warming increased gross ecosystem productivity, the higher plant biomass did not compensate for the much greater increase in C losses. Ecosystem respiration and net ecosystem CO2 losses were significantly higher in warmed plots compared to control ones. We suggest that this was due to increased soil respiration, since soil carbon pools were lower in warmed soils, at least in the upper horizons. Our study thus supports the general hypothesis that tundra ecosystems turn from a carbon sink to a carbon source when temperatures increase in the short-term. Since lichens, which produce low quality litter, increased their biomass significantly with warming in this specific ecosystem type, CO2 losses may slow down in the long-term.
NH4+ ad-/desorption in sequencing batch reactors: simulation, laboratory and full-scale studies
2008 - Water Sci. Technol., 58: 345-350
Abstract:
Significant NH4-N balance deficits were found during the measurement campaigns for the data collection for dynamic simulation studies at five full-scale sequencing batch reactor (SBR) waste water treatment plants (WWTPs), as well as during subsequent calibrations at the investigated plants. Subsequent lab scale investigations showed high evidence for dynamic, cycle-specific NH4+ ad-/desorption to the activated flocs as one reason for this balance deficit. This specific dynamic was investigated at five full-scale SBR plants for the search of the general causing mechanisms. The general mechanism found was a NH4+ desorption from the activated flocs at the end of the nitrification phase with subsequent nitrification and a chemical NH4+ adsorption at the flocs in the course of the filling phases. This NH4+ ad-/desorption corresponds to an antiparallel K+ ad/-desorption.One reasonable full-scale application was investigated at three SBR plants, a controlled filling phase at the beginning of the sedimentation phase. The results indicate that this kind of filling event must be specifically hydraulic controlled and optimised in order to prevent too high waste water break through into the clear water phase, which will subsequently be discarded.
Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH
2008 - Appl. Environ. Microbiol., 74: 5068-5077
Abstract:
Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes is a widely used method to detect and quantify microorganisms by fluorescence microscopy in environmental samples and medical specimens. Difficulties with FISH arise if the rRNA content of the probe-target organisms is low, causing dim fluorescence signals that are not detectable against background fluorescence. This limitation is ameliorated by technical modifications such as catalysed reporter deposition-fluorescence in situ hybridization (CARD-FISH), but to date the minimal rRNA copy numbers needed to obtain a visible signal of a microbial cell after FISH or CARD-FISH have not been determined. In this study, a novel competitive FISH approach was developed and used to quantify, based on a thermodynamic model of probe competition, the 16S rRNA copy numbers per cell required to detect bacteria by FISH and CARD-FISH with oligonucleotide probes in mixed pure cultures and in activated sludge. The detection limit of conventional FISH with Cy3-labelled probe EUB338-I was found to be 370±45 16S rRNA molecules per cell for E. coli hybridized on microscope glass slides, and 1,400±170 16S rRNA copies per E. coli cell in activated sludge. For CARD-FISH the respective values ranged from 8.9±1.5 to 14±2 and from 36±6 to 54±7 16S rRNA molecules per cell, indicating that the sensitivity of CARD-FISH was 26 to 41-fold higher than that of conventional FISH. These results suggest that optimized FISH protocols using oligonucleotide probes could be suitable for more recent applications of FISH, for example to detect mRNA in situ in microbial cells.
Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii"
2008 - Environ. Microbiol., 10: 3043-3056
Abstract:
Nitrite-oxidizing bacteria of the genus Nitrospira are ubiquitous in natural ecosystems and also in wastewater treatment plants. Nitrospira are members of a distinct phylum, not closely related to other nitrifiers, and no genomic sequences from this genus have been available so far. Here we applied an environmental genomics approach to sequence and assemble a 137 kbp-long genome fragment of 'Candidatus Nitrospira defluvii', which had been enriched from activated sludge and belongs to Nitrospira sublineage I without isolated representatives. The annotation of this contig, which carried the 16S rRNA gene of N. defluvii, offered first insight into the genome of Nitrospira. Surprisingly, we found a gene similar to genes encoding chlorite dismutase (CLD), an enzyme degrading chlorite (ClO(2)(-)) to Cl(-) and O(2). To date, CLDs with high catalytic activity have been found only in perchlorate- and chlorate-reducing bacteria but not in nitrifiers. Heterologous expression in E. coli followed by enzymatic tests confirmed that this gene of Nitrospira encodes a highly active CLD, which is also expressed in situ by Nitrospira, indicating that this nitrite oxidizer might be involved in the bioremediation of perchlorate and chlorite. Phylogenetic analyses showed that CLD and related proteins are widely distributed among the Bacteria and Archaea, and indicated that this enzyme family appeared relatively early in evolution, has been subject to functional diversification and might play yet unknown roles in microbial metabolism.
Nitrification in terrestrial hot springs of Iceland and Kamchatka
2008 - FEMS Microbiol. Ecol., 64: 167-174
Abstract:
Archaea have been detected recently as a major and often dominant component of the microbial communities performing ammonia oxidation in terrestrial and marine environments. In a molecular survey of archaeal ammonia monooxygenase (AMO) genes in terrestrial hot springs of Iceland and Kamchatka, the amoA gene encoding the alpha-subunit of AMO was detected in a total of 14 hot springs out of the 22 investigated. Most of these amoA-positive hot springs had temperatures between 82 and 97 degrees C and pH range between 2.5 and 7. In phylogenetic analyses, these amoA genes formed three independent lineages within the known sequence clusters of marine or soil origin. Furthermore, in situ gross nitrification rates in Icelandic hot springs were estimated by the pool dilution technique directly on site. At temperatures above 80 degrees C, between 56 and 159 mumol NO(3)(-) L(-1) mud per day was produced. Furthermore, addition of ammonium to the hot spring samples before incubation yielded a more than twofold higher potential nitrification rate, indicating that the process was limited by ammonia supply. Our data provide evidence for an active role of archaea in nitrification of hot springs in a wide range of pH values and at a high temperature.
A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring
2008 - Proc. Natl. Acad. Sci. USA, 105: 2134-2139
Abstract:
The recent discovery of ammonia-oxidizing archaea (AOA) dramatically changed our perception of the diversity and evolutionary history of microbes involved in nitrification. In this study, a moderately thermophilic (46°C) ammonia-oxidizing enrichment culture, which had been seeded with biomass from a hot spring, was screened for ammonia oxidizers. Although gene sequences for crenarchaeotal 16S rRNA and two subunits of the ammonia monooxygenase (amoA and amoB) were detected via PCR, no hints for known ammonia-oxidizing bacteria were obtained. Comparative sequence analyses of these gene fragments demonstrated the presence of a single operational taxonomic unit and thus enabled the assignment of the amoA and amoB sequences to the respective 16S rRNA phylotype, which belongs to the widely distributed group I.1b (soil group) of the Crenarchaeota. Catalyzed reporter deposition (CARD)–FISH combined with microautoradiography (MAR) demonstrated metabolic activity of this archaeon in the presence of ammonium. This finding was corroborated by the detection of amoA gene transcripts in the enrichment. CARD-FISH/MAR showed that the moderately thermophilic AOA is highly active at 0.14 and 0.79 mM ammonium and is partially inhibited by a concentration of 3.08 mM. The enriched AOA, which is provisionally classified as "Candidatus Nitrososphaera gargensis," is the first described thermophilic ammonia oxidizer and the first member of the crenarchaeotal group I.1b for which ammonium oxidation has been verified on a cellular level. Its preference for thermophilic conditions reinvigorates the debate on the thermophilic ancestry of AOA.
Physiological and phylogenetical characterization of a new lithoautotrophic nitrite-oxidizing bacterium 'Candidatus Nitrospira bockiana' sp. nov
2008 - Int. J. Syst. Evol. Microbiol., 58: 242-250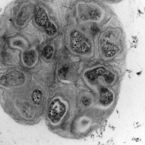
Abstract:
A new isolate of a lithoautotrophic nitrite-oxidizing bacterium was obtained from internal corrosion deposits from a steel pipeline of the Moscow heating system. The organism oxidized nitrite as the sole energy source and fixed carbon dioxide as the only carbon source. The cells were extremely pleomorphic: loosely wound spirals, slightly curved and even straight rods were detected, as well as coccoid cells. The highest rate of nitrite consumption (1.5 mM nitrite as substrate) was measured at 42 degrees C, with a temperature range of 28-44 degrees C. In enrichment cultures with Nocardioides sp. as an accompanying organism, optimal oxidation of 5.8 mM nitrite occurred at 45 degrees C, with a range of 28-48 degrees C. Neither pyruvate nor yeast extract stimulated nitrification. Organotrophic growth was not observed. Phylogenetic analysis of 16S rRNA gene sequences revealed that the novel isolate represents a new sublineage of the genus Nitrospira. On the basis of physiological, chemotaxonomic and molecular characteristics, the name 'Candidatus Nitrospira bockiana' is proposed.
Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function
2007 - Environ Microbiol., 9: 1878-1889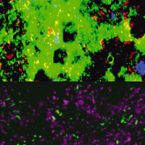
Abstract:
We have coupled fluorescence in situ hybridization (FISH) with Raman microscopy for simultaneous cultivation-independent identification and determination of (13)C incorporation into microbial cells. Highly resolved Raman confocal spectra were generated for individual cells which were grown in minimal medium where the ratio of (13)C to (12)C content of the sole carbon source was incrementally varied. Cells which were (13)C-labelled through anabolic incorporation of the isotope exhibited key red-shifted spectral peaks, the calculated 'red shift ratio' (RSR) being highly correlated with the (13)C-content of the cells. Subsequently, Raman instrumentation and FISH protocols were optimized to allow combined epifluorescence and Raman imaging of Fluos, Cy3 and Cy5-labelled microbial populations at the single cell level. Cellular (13)C-content determinations exhibited good congruence between fresh cells and FISH hybridized cells indicating that spectral peaks, including phenylalanine resonance, which were used to determine (13)C-labelling, were preserved during fixation and hybridization. In order to demonstrate the suitability of this technology for structure-function analyses in complex microbial communities, Raman-FISH was deployed to show the importance of Pseudomonas populations during naphthalene degradation in groundwater microcosms. Raman-FISH extends and complements current technologies such as FISH-microautoradiography and stable isotope probing in that it can be applied at the resolution of single cells in complex communities, is quantitative if suitable calibrations are performed, can be used with stable isotopes and has analysis times of typically 1 min per cell.
Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis
2007 - Appl. Microbiol. Biotech., 75: 237-248
Abstract:
Traditional cultivation-based methods to quantify microbial abundance are not suitable for analyses of microbial communities in environmental or medical samples, which consist mainly of uncultured microorganisms. Recently, different cultivation-independent quantification approaches have been developed to overcome this problem. Some of these techniques use specific fluorescence markers, for example ribosomal ribonucleic acid targeted oligonucleotide probes, to label the respective target organisms. Subsequently, the detected cells are visualized by fluorescence microscopy and are quantified by direct visual cell counting or by digital image analysis. This article provides an overview of these methods and some of their applications with emphasis on (semi-)automated image analysis solutions.
Microbial landscapes: new paths to biofilm research
2007 - Nature Rev. Microbiol., 5: 76-81
Abstract:
It is the best of times for biofilm research. Systems biology approaches are providing new insights into the genetic regulation of microbial functions, and sophisticated modelling techniques are enabling the prediction of microbial community structures. Yet it is also clear that there is a need for ecological theory to contribute to our understanding of biofilms. Here, we suggest a concept for biofilm research that is spatially explicit and solidly rooted in ecological theory, which might serve as a universal approach to the study of the numerous facets of biofilms.
Soil carbon and nitrogen dynamics along a latitudinal transect in Western Siberia, Russia
2006 - Biogeochemistry, 81: 239-252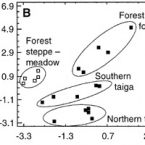
Abstract:
An 1800-km South to North transect (N 53°43′ to 69°43′) through Western Siberia was established to study the interaction of nitrogen and carbon cycles. The transect comprised all major vegetation zones from steppe, through taiga to tundra and corresponded to a natural temperature gradient of 9.5°C mean annual temperature (MAT). In order to elucidate changes in the control of C and N cycling along this transect, we analyzed physical and chemical properties of soils and microbial structure and activity in the organic and in the mineral horizons, respectively. The impact of vegetation and climate exerted major controls on soil C and N pools (e.g., soil organic matter, total C and dissolved inorganic nitrogen) and process rates (gross N mineralization and heterotrophic respiration) in the organic horizons. In the mineral horizons, however, the impact of climate and vegetation was less pronounced. Gross N mineralization rates decreased in the organic horizons from south to north, while remaining nearly constant in the mineral horizons. Especially, in the northern taiga and southern tundra gross nitrogen mineralization rates were higher in the mineral compared to organic horizons, pointing to strong N limitation in these biomes. Heterotrophic respiration rates did not exhibit a clear trend along the transect, but were generally higher in the organic horizon compared to mineral horizons. Therefore, C and N mineralization were spatially decoupled at the northern taiga and tundra. The climate change implications of these findings (specifically for the Arctic) are discussed.
Wastewater treatment: a model system for microbial ecology
2006 - Trends Biotechnol., 24: 483-489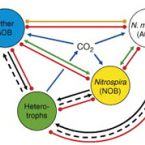
Abstract:
Biological wastewater treatment is among the most important biotechnological applications and, as drivers of the key processes, microorganisms are central to its success. Therefore, the study of wastewater microorganisms has obvious applied significance; however, the importance of wastewater treatment reactors as model systems for microbial ecology is often overlooked. Modern molecular techniques, including environmental genomics, have identified unexpected microbial key players for nutrient removal and sludge bulking and/or foaming, and provided many exciting insights into the diversity, functions and niche differentiations of these predominantly uncultivated microorganisms. It is now time for wastewater microbiology to be recognized as a mature and dynamic discipline in its own right, offering much toward a deeper understanding of life in complex microbial communities. Here, we consider selected key findings to illustrate the past and future roles of molecular ecophysiology and genomics in the development of wastewater microbiology as an important subdiscipline of microbial ecology.
Ecophysiology and niche differentiation of Nitrospira-like bacteria, the key nitrite oxidizers in wastewater treatment plants
2006 - Water Sci. Tech., 54: 21-27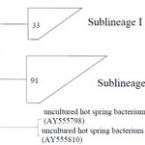
Abstract:
Nitrite-oxidizing bacteria of the genus Nitrospira are key nitrifiers in wastewater treatment plants. Pure cultures of these organisms are unavailable, but cultivation-independent molecular methods make it possible to detect Nitrospira-like bacteria in environmental samples and to investigate their ecophysiology. Comprehensive screening of natural and engineered habitats and of public databases for 16S rRNA sequences of Nitrospira-like bacteria revealed a surprisingly high biodiversity in the genus Nitrospira, which comprises at least four phylogenetic sublineages. All Nitrospira-like bacteria detected in wastewater treatment plants belonged to the sublineages I and II. Subsequently, the population dynamics of different Nitrospira-like bacteria were monitored, by quantitative fluorescence in situ hybridization with rRNA-targeted probes, confocal laser scanning microscopy and digital image analysis, during incubation of nitrifying activated sludge in media containing different nitrite concentrations. These experiments showed that Nitrospira-like bacteria, which were affiliated with the phylogenetic sublineages I or II of the genus Nitrospira, responded differently to nitrite concentration shifts. Previously unknown properties of Nitrospira-like bacteria were discovered in the course of an environmental genomics project. Implications of the obtained results for fundamental understanding of the microbial ecology of nitrite oxidizers as well as for future improvement of nutrient removal in wastewater treatment plants are discussed.
Nitrite concentration influences the population structure of Nitrospira-like bacteria
2006 - Environ. Microbiol., 8: 1487-1495
Abstract:
Chemolithoautotrophic nitrite oxidizers of the genus Nitrospira are a monophyletic but diverse group of organisms, are widely distributed in many natural habitats, and play a key role in nitrogen elimination during biological wastewater treatment. Phylogenetic analyses of cloned 16S rRNA genes and fluorescence in situ hybridization with newly developed rRNA-targeted oligonucleotide probes revealed coexistence of uncultured members of sublineages I and II of the genus Nitrospira in biofilm and activated sludge samples taken from nitrifying wastewater treatment plants. Quantitative microscopic analyses of their spatial arrangement relative to ammonia oxidizers in the biofilm and activated sludge flocs showed that members of the Nitrospira sublineage I occurred significantly more often in immediate vicinity to ammonia oxidizers than would be expected from random community assembly while such a relationship was not observed for Nitrospira sublineage II. This spatial distribution suggested a niche differentiation of these coexisting Nitrospira populations with respect to their preferred concentrations of nitrite. This hypothesis was tested by mathematical modelling of nitrite consumption and resulting nitrite gradients in nitrifying biofilms and by quantifying the abundance of sublineage I and II Nitrospira in activated sludge during incubations with nitrite in different concentrations. Consistent with the observed localization patterns, a higher nitrite concentration selected for sublineage I but suppressed sublineage II Nitrospira.
Deciphering the evolution and metabolism of an anammox bacterium from a community genome
2006 - Nature, 440: 790-794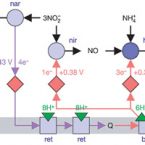
Abstract:
Anaerobic ammonium oxidation (anammox) has become a main focus in oceanography and wastewater treatment. It is also the nitrogen cycle's major remaining biochemical enigma. Among its features, the occurrence of hydrazine as a free intermediate of catabolism, the biosynthesis of ladderane lipids and the role of cytoplasm differentiation are unique in biology. Here we use environmental genomics the reconstruction of genomic data directly from the environment to assemble the genome of the uncultured anammox bacterium Kuenenia stuttgartiensis from a complex bioreactor community. The genome data illuminate the evolutionary history of the Planctomycetes and allow us to expose the genetic blueprint of the organism's special properties. Most significantly, we identified candidate genes responsible for ladderane biosynthesis and biological hydrazine metabolism, and discovered unexpected metabolic versatility.
Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge
2006 - Environ. Microbiol., 8: 405-415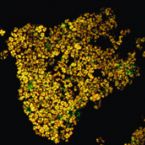
Abstract:
Previously uncultured nitrite-oxidizing bacteria affiliated to the genus Nitrospira have for the first time been successfully enriched from activated sludge from a municipal wastewater treatment plant. During the enrichment procedure, the abundance of the Nitrospira-like bacteria increased to approximately 86% of the total bacterial population. This high degree of purification was achieved by a novel enrichment protocol, which exploits physiological features of Nitrospira-like bacteria and includes the selective repression of coexisting Nitrobacter cells and heterotrophic contaminants by application of ampicillin in a final concentration of 50 microg ml(-1). The enrichment process was monitored by electron microscopy, fluorescence in situ hybridization (FISH) with rRNA-targeted probes and fatty acid profiling. Phylogenetic analysis of 16S rRNA gene sequences revealed that the enriched bacteria represent a novel Nitrospira species closely related to uncultured Nitrospira-like bacteria previously found in wastewater treatment plants and nitrifying bioreactors. The enriched strain is provisionally classified as 'Candidatus Nitrospira defluvii'.
Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase
2006 - Proc. Natl. Acad. Sci. USA, 7: 2363-2367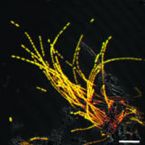
Abstract:
135 years ago Ferdinand Cohn, the founder of bacteriology, microscopically observed a conspicuous filamentous bacterium with a complex life cycle and described it as Crenothrix polyspora. This uncultured bacterium is infamous for mass developments in drinking water systems, but its phylogeny and physiology remained unknown. We show that C. polyspora is a gammaproteobacterium closely related to methanotrophs and capable of oxidizing methane. We discovered that C. polyspora encodes a phylogenetically very unusual particulate methane monooxygenase whose expression is strongly increased in the presence of methane. Our findings demonstrate a previously unrecognized complexity of the evolutionary history and cell biology of methane-oxidizing bacteria.
daime, a novel image analysis program for microbial ecology and biofilm research
2006 - Environ. Microbiol., 8: 200-213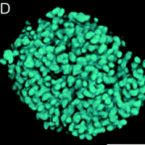
Abstract:
Combinations of microscopy and molecular techniques to detect, identify and characterize microorganisms in environmental and medical samples are widely used in microbial ecology and biofilm research. The scope of these methods, which include fluorescence in situ hybridization (FISH) with rRNA-targeted probes, is extended by digital image analysis routines that extract from micrographs important quantitative data. Here we introduce daime (digital image analysis in microbial ecology), a new computer program integrating 2-D and 3-D image analysis and visualization functionality, which has previously not been available in a single open-source software package. For example, daime automatically finds 2-D and 3-D objects in images and confocal image stacks, and offers special functions for quantifying microbial populations and evaluating new FISH probes. A novel feature is the quantification of spatial localization patterns of microorganisms in complex samples like biofilms. In combination with '3D-FISH', which preserves the 3-D structure of samples, this stereological technique was applied in a proof of principle experiment on activated sludge and provided quantitative evidence that functionally linked ammonia and nitrite oxidizers cluster together in their habitat. This image analysis method complements recent molecular techniques for analysing structure-function relationships in microbial communities and will help to characterize symbiotic interactions among microorganisms.
Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays
2006 - Curr. Opin. Biotechnol., 17: 1-9
Abstract:
The ecophysiology of microorganisms has been at the heart of microbial ecology since its early days, but only during the past decade have methods become available for cultivation-independent, direct identification of microorganisms in complex communities and for the simultaneous investigation of their activity and substrate uptake patterns. The combination of fluorescence in situ hybridization (FISH) and microautoradiography (MAR) is currently the most widely applied tool for revealing physiological properties of microorganisms in their natural environment with single-cell resolution. For example, this technique has been used in wastewater treatment and marine systems to describe the functional properties of newly discovered species, and to identify microorganisms responsible for key physiological processes. Recently, the scope of FISH-MAR was extended by rendering it quantitative and by combining it with microelectrode measurements or stable isotope probing. Isotope arrays have also been developed that exploit the parallel detection offered by DNA microarrays to measure incorporation of labelled substrate into the rRNA of many community members in a single experiment.
Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-micrautoradiography to study a methanol-fed denitrifying microbial community
2004 - Appl. Environ. Microbiol., 70: 588-596
Abstract:
A denitrifying microbial consortium was enriched in an anoxically operated, methanol-fed sequencing batch reactor (SBR) fed with a mineral salts medium containing methanol as the sole carbon source and nitrate as the electron acceptor. The SBR was inoculated with sludge from a biological nutrient removal activated sludge plant exhibiting good denitrification. The SBR denitrification rate improved from less than 0.02 mg of NO(3)(-)-N mg of mixed-liquor volatile suspended solids (MLVSS)(-1) h(-1) to a steady-state value of 0.06 mg of NO(3)(-)-N mg of MLVSS(-1) h(-1) over a 7-month operational period. At this time, the enriched microbial community was subjected to stable-isotope probing (SIP) with [(13)C]methanol to biomark the DNA of the denitrifiers. The extracted [(13)C]DNA and [(12)C]DNA from the SIP experiment were separately subjected to full-cycle rRNA analysis. The dominant 16S rRNA gene phylotype (group A clones) in the [(13)C]DNA clone library was closely related to those of the obligate methylotrophs Methylobacillus and Methylophilus in the order Methylophilales of the Betaproteobacteria (96 to 97% sequence identities), while the most abundant clone groups in the [(12)C]DNA clone library mostly belonged to the family Saprospiraceae in the Bacteroidetes phylum. Oligonucleotide probes for use in fluorescence in situ hybridization (FISH) were designed to specifically target the group A clones and Methylophilales (probes DEN67 and MET1216, respectively) and the Saprospiraceae clones (probe SAP553). Application of these probes to the SBR biomass over the enrichment period demonstrated a strong correlation between the level of SBR denitrification and relative abundance of DEN67-targeted bacteria in the SBR community. By contrast, there was no correlation between the denitrification rate and the relative abundances of the well-known denitrifying genera Hyphomicrobium and Paracoccus or the Saprospiraceae clones visualized by FISH in the SBR biomass. FISH combined with microautoradiography independently confirmed that the DEN67-targeted cells were the dominant bacterial group capable of anoxic [(14)C]methanol uptake in the enriched biomass. The well-known denitrification lag period in the methanol-fed SBR was shown to coincide with a lag phase in growth of the DEN67-targeted denitrifying population. We conclude that Methylophilales bacteria are the dominant denitrifiers in our SBR system and likely are important denitrifiers in full-scale methanol-fed denitrifying sludges.
Fluorescence in situ hybridisation for the identification and charcterisation of prokaryotes
2003 - Curr. Opin. Microbiol., 6: 302-309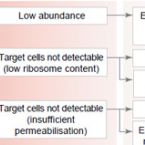
Abstract:
Fluorescence in situ hybridisation with rRNA-targeted nucleic acid probes can be used to directly identify microorganisms within complex samples in a few hours and therefore has widespread application in environmental and medical microbiology. The past year has seen significant methodological improvements in fluorescence in situ hybridisation, as well as in the combination of this method with other techniques for inferring functional traits of microorganisms within their environment.
Microbial community composition and function in wastewater treatment plants
2002 - Antonie van Leeuwenhoek, 81: 665-680
Abstract:
Biological wastewater treatment has been applied for more than a century to ameliorate anthropogenic damage to the environment. But only during the last decade the use of molecular tools allowed to accurately determine the composition, and dynamics of activated sludge and biofilm microbial communities. Novel, in many cases yet not cultured bacteria were identified to be responsible for filamentous bulking and foaming as well as phosphorus and nitrogen removal in these systems. Now, methods are developed to infer the in situ physiology of these bacteria. Here we provide an overview of what is currently known about the identity and physiology of some of the microbial key players in activated sludge and biofilm systems.
Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization
2001 - Appl. Environ. Microbiol., 67: 5810-5818
Abstract:
Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes has found widespread application for analyzing the composition of microbial communities in complex environmental samples. Although bacteria can quickly be detected by FISH, a reliable method to determine absolute numbers of FISH-stained cells in aggregates or biofilms has, to our knowledge, never been published. In this study we developed a semiautomated protocol to measure the concentration of bacteria (in cells per volume) in environmental samples by a combination of FISH, confocal laser scanning microscopy, and digital image analysis. The quantification is based on an internal standard, which is introduced by spiking the samples with known amounts of Escherichia coli cells. This method was initially tested with artificial mixtures of bacterial cultures and subsequently used to determine the concentration of ammonia-oxidizing bacteria in a municipal nitrifying activated sludge. The total number of ammonia oxidizers was found to be 9.8 x 10(7) +/- 1.9 x 10(7) cells ml(-1). Based on this value, the average in situ activity was calculated to be 2.3 fmol of ammonia converted to nitrite per ammonia oxidizer cell per h. This activity is within the previously determined range of activities measured with ammonia oxidizer pure cultures, demonstrating the utility of this quantification method for enumerating bacteria in samples in which cells are not homogeneously distributed.
In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in waste water treatment plants
2001 - Appl. Environ. Microbiol., 67: 5273-5284
Abstract:
Uncultivated Nitrospira-like bacteria in different biofilm and activated-sludge samples were investigated by cultivation-independent molecular approaches. Initially, the phylogenetic affiliation of Nitrospira-like bacteria in a nitrifying biofilm was determined by 16S rRNA gene sequence analysis. Subsequently, a phylogenetic consensus tree of the Nitrospira phylum including all publicly available sequences was constructed. This analysis revealed that the genus Nitrospira consists of at least four distinct sublineages. Based on these data, two 16S rRNA-directed oligonucleotide probes specific for the phylum and genus Nitrospira, respectively, were developed and evaluated for suitability for fluorescence in situ hybridization (FISH). The probes were used to investigate the in situ architecture of cell aggregates of Nitrospira-like nitrite oxidizers in wastewater treatment plants by FISH, confocal laser scanning microscopy, and computer-aided three-dimensional visualization. Cavities and a network of cell-free channels inside the Nitrospira microcolonies were detected that were water permeable, as demonstrated by fluorescein staining. The uptake of different carbon sources by Nitrospira-like bacteria within their natural habitat under different incubation conditions was studied by combined FISH and microautoradiography. Under aerobic conditions, the Nitrospira-like bacteria in bioreactor samples took up inorganic carbon (as HCO(3)(-) or as CO(2)) and pyruvate but not acetate, butyrate, and propionate, suggesting that these bacteria can grow mixotrophically in the presence of pyruvate. In contrast, no uptake by the Nitrospira-like bacteria of any of the carbon sources tested was observed under anoxic or anaerobic conditions.
Nitrification in sequencing biofilm batch reactors: lessons from molecular approaches
2001 - Water Sci. Technol., 43: 9-18
Abstract:
The nitrifying microbial diversity and population structure of a sequencing biofilm batch reactor receiving sewage with high ammonia and salt concentrations (SBBR 1) was analyzed by the full-cycle rRNA approach. The diversity of ammonia-oxidizers in this reactor was additionally investigated using comparative sequence analysis of a gene fragment of the ammonia monooxygenase (amoA), which represents a key enzyme of all ammonia-oxidizers. Despite the "extreme" conditions in the reactor, a surprisingly high diversity of ammonia- and nitrite-oxidizers was observed to occur within the biofilm. In addition, molecular evidence for the existence of novel ammonia-oxidizers is presented. Quantification of ammonia- and nitrite-oxidizers in the biofilm by Fluorescent In situ Hybridization (FISH) and digital image analysis revealed that ammonia-oxidizers occurred in higher cell numbers and occupied a considerably larger share of the total biovolume than nitrite-oxidizing bacteria. In addition, ammonia oxidation rates per cell were calculated for different WWTPs following the quantification of ammonia-oxidizers by competitive PCR of an amoA gene fragment. The morphology of nitrite-oxidizing, unculturable Nitrospira-like bacteria was studied using FISH, confocal laser scanning microscopy (CLSM) and three-dimensional visualization. Thereby, a complex network of microchannels and cavities was detected within microcolonies of Nitrospira-like bacteria. Microautoradiography combined with FISH was applied to investigate the ability of these organisms to use CO2 as carbon source and to take up other organic substrates under varying conditions. Implications of the obtained results for fundamental understanding of the microbial ecology of nitrifiers as well as for future improvement of nutrient removal in wastewater treatment plants (WWTPs) are discussed.
Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: Diversity and in situ physiology
2000 - Water Sci. Technol., 41: 85-90
Probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set
1999 - Syst. Appl. Microbiol., 22: 434-444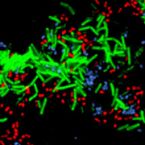
Abstract:
In situ hybridization with rRNA-targeted oligonucleotide probes has become a widely applied tool for direct analysis of microbial population structures of complex natural and engineered systems. In such studies probe EUB338 (AMANN et al., 1990) is routinely used to quantify members of the domain Bacteria with a sufficiently high cellular ribosome content. Recent reevaluations of probe EUB338 coverage based on all publicly available 16S rRNA sequences, however, indicated that important bacterial phyla, most notably the Planctomycetales and Verrucomicrobia, are missed by this probe. We therefore designed and evaluated two supplementary versions (EUB338-II and EUB338-III) of probe EUB338 for in situ detection of most of those phyla not detected with probe EUB338. In situ dissociation curves with target and non-target organisms were recorded under increasing stringency to optimize hybridization conditions. For that purpose a digital image software routine was developed. In situ hybridization of a complex biofilm community with the three EUB338 probes demonstrated the presence of significant numbers of probe EUB338-II and EUB338-III target organisms. The application of EUB338, EUB338-II and EUB338-III should allow a more accurate quantification of members of the domain Bacteria in future molecular ecological studies.