Publications
Publications in peer reviewed journals
Genome-guided design of a novel defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium
2016 - Nature Microbiol, 2: 16215
Abstract:
Protection against enteric infections, also termed colonization resistance, results from mutualistic interactions of the host and its indigenous microbes. The gut microbiota of humans and mice is highly diverse and it is therefore challenging to assign specific properties to its individual members. Here, we have used a collection of murine bacterial strains and a modular design approach to create a minimal bacterial community that, once established in germ-free mice, provided colonization resistance against the human enteric pathogen Salmonella enterica serovar Typhimurium (S. Tm). Initially, a community of 12 strains, termed Oligo-Mouse Microbiota (Oligo-MM12), representing members of the major bacterial phyla in the murine gut, was selected. This community was stable over consecutive mouse generations and provided colonization resistance against S. Tm infection, albeit not to the degree of a conventional complex microbiota. Comparative (meta)genome analyses identified functions represented in a conventional microbiome but absent from the Oligo-MM12. By genome-informed design, we created an improved version of the Oligo-MM community harbouring three facultative anaerobic bacteria from the Mouse Intestinal Bacterial Collection (miBC) that provided conventional-like colonization resistance. In conclusion, we have established a highly versatile experimental system that showed efficacy in an enteric infection model. Thus, in combination with exhaustive bacterial strain collections and systems-based approaches, genomeguided design can be used to generate insights into microbe–microbe and microbe–host interactions for the investigation of ecological and disease-relevant mechanisms in the intestine.
Environmental enteric dysfunction and growth failure/stunting in global child health
2016 - Pediatrics, 138: e20160641
Abstract:
Approximately 25% of the world’s children under age 5 years have stunted growth, which is associated with increased mortality, cognitive dysfunction, and loss of productivity. Reducing by 40% the number of stunted children is a global target for 2030. The pathogenesis of stunting is poorly understood. Pre- and post-natal nutritional deficits and enteric and systemic infections clearly contribute, but recent findings implicate a central role for environmental enteric dysfunction (EED), a generalized disturbance of small intestinal structure and function found at high prevalence in children living under unsanitary conditions. Mechanisms contributing to growth failure in EED include intestinal leakiness and heightened permeability, gut inflammation, dysbiosis and bacterial translocation, systemic inflammation, and nutrient malabsorption. Since EED has multiple causal pathways, approaches to manage it need to be multi-faceted. Potential interventions to tackle EED include: a) reduction of exposure to feces and contact with animals through programs like improved water, sanitation and hygiene (WASH); b) breastfeeding and enhanced dietary diversity; c) probiotics and prebiotics; d) nutrient supplements including zinc, polyunsaturated fatty acids, and amino acids; e) anti-inflammatory agents such as 5-aminosalicyclic acid; and f) antibiotics in the context of acute malnutrition and infection. Better understanding of the underlying causes of EED, and development of non-invasive, practical, simple, and affordable point of care diagnostic tools remain key gaps. ‘Omics’ technologies (genomics, epigenomics, transcriptomics, proteomics, and metabolomics) and stable isotope techniques (e.g., 13Carbon breath tests) targeted at children and their intestinal microbiota will enhance our ability to successfully identify, manage, and prevent the disorder.
Bacterial nutrient foraging in a mouse model of enteral nutrient deprivation: Insight into the gut origin of sepsis
2016 - Am J Physiol Gastrointest Liver Physiol, 311: G734-G743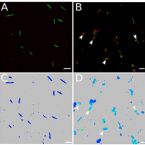
Abstract:
Total parenteral nutrition (TPN) leads to a shift in small intestinal microbiota with a characteristic dominance of Proteobacteria. This study examined how metabolomic changes within the small bowel support an altered microbial community in enterally deprived mice.
C57BL/6 mice were given TPN or enteral chow. Metabolomic analysis of jejunal contents was performed by liquid chromatography/mass spectrometry (LC/MS). In some experiments, leucine in TPN was partly substituted with (13)C-leucine. Additionally, jejunal contents from TPN dependent and enterally fed mice were gavaged into germ-free mice to reveal if the TPN phenotype was transferrable.
Small bowel contents of TPN mice maintained an amino acid composition similar to that of the TPN solution. Mass spectrometry analysis of small bowel contents of TPN dependent mice showed increased concentration of (13)C compared to fed mice receiving saline enriched with (13)C-leucine. (13)C-leucine added to the serosal side of Ussing chambers showed rapid permeation across TPN-dependent jejunum, suggesting increased transmucosal passage. Single-cell analysis by fluorescence in situ hybridization (FISH) - NanoSIMS demonstrated uptake of (13)C-leucine by TPN-associated bacteria, with preferential uptake by Enterobacteriaceae. Gavage of small bowel effluent from TPN mice into germ-free, fed mice resulted in a trend toward the pro-inflammatory TPN-phenotype with loss of epithelial barrier function.
TPN-dependence leads to increased permeation of TPN-derived nutrients into the small intestinal lumen, where they are predominately utilized by Enterobacteriaceae. The altered metabolomic composition of the intestinal lumen during TPN promotes dysbiosis.Phylogenetic and genomic analysis of Methanomassiliicoccales in wetlands and animal intestinal tracts reveals clade-specific habitat preferences
2016 - FEMS Microbiol Ecol, 92: fiv149
Abstract:
Methanogenic Thermoplasmata of the novel order Methanomassiliicoccales were recently discovered in human and animal gastro-intestinal tracts (GITs). However their distribution in other methanogenic environments has not been addressed systematically. Here we surveyed Methanomassiliicoccales presence in wetland soils, a globally important source of methane emissions to the atmosphere, and in the GITs of different animals by PCR targeting their 16S rRNA and methyl:coenzyme M reductase (α-subunit) genes. We detected Methanomassiliicoccales in all 16 peat soils investigated, indicating their wide distribution in these habitats. Additionally, we detected their genes in various animal feces. Methanomassiliicoccales were subdivided in two broad phylogenetic clades designated 'environmental' and 'GIT' clades based on differential, although non-exclusive, habitat preferences of their members. A well-supported cluster within the environmental clade comprised more than 80% of all wetland 16S rRNA gene sequences. Metagenome assembly from bovine rumen fluid enrichments resulted in two almost complete genomes of both Methanomassiliicoccales clades. Comparative genomics revealed that members of the environmental clade contain larger genomes and a higher number of genes encoding anti-oxidative enzymes than animal GIT clade representatives. This study highlights the wide distribution of Methanomassiliicoccales in wetlands, which suggests that they contribute to methane emissions from these climate-relevant ecosystems.