Publications
Publications in peer reviewed journals
Prediction of microbial phenotypes based on comparative genomics
2015 - BMC Bioinformatics, 16: S1
Abstract:
The accessibility of almost complete genome sequences of uncultivable microbial species from metagenomes necessitates computational methods predicting microbial phenotypes solely based on genomic data. Here we investigate how comparative genomics can be utilized for the prediction of microbial phenotypes. The PICA framework facilitates application and comparison of different machine learning techniques for phenotypic trait prediction. We have improved and extended PICA's support vector machine plug-in and suggest its applicability to large-scale genome databases and incomplete genome sequences
Conductive consortia
2015 - Nature News & Views, 526: 513-514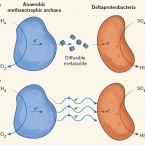
Abstract:
Physiological analyses, electron microscopy and single-cell chemical imaging suggest that direct electron transfer occurs between the members of methane-oxidizing microbial consortia.
Advancements in the application of NanoSIMS and Raman microspectroscopy to investigate the activity of microbial cells in soils
2015 - FEMS Microbiology Ecology - *Editor's Choice Article*, in press
Abstract:
The combined approach of incubating environmental samples with stable isotope-labeled substrates followed by single-cell analyses through high-resolution secondary ion mass spectrometry (NanoSIMS) or Raman microspectroscopy provides insights into the in situ function of microorganisms. This approach has found limited application in soils presumably due to the dispersal of microbial cells in a large background of particles. We developed a pipeline for the efficient preparation of cell extracts from soils for subsequent single-cell methods by combining cell detachment with separation of cells and soil particles followed by cell concentration. The procedure was evaluated by examining its influence on cell recoveries and microbial community composition across two soils. This approach generated a cell fraction with considerably reduced soil particle load and of sufficient small size to allow single-cell analysis by NanoSIMS, as shown when detecting active N2-fixing and cellulose-responsive microorganisms via 15N2 and 13C-UL-cellulose incubations, respectively. The same procedure was also applicable for Raman microspectroscopic analyses of soil microorganisms, assessed via microcosm incubations with a 13C-labeled carbon source and deuterium oxide (D2O, a general activity marker). The described sample preparation procedure enables single-cell analysis of soil microorganisms using NanoSIMS and Raman microspectroscopy, but should also facilitate single-cell sorting and sequencing.
A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes.
2015 - Front Microbiol, 6: 731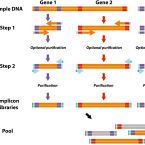
Abstract:
High throughput sequencing of phylogenetic and functional gene amplicons provides tremendous insight into the structure and functional potential of complex microbial communities. Here, we introduce a highly adaptable and economical PCR approach to barcoding and pooling libraries of numerous target genes. In this approach, we replace gene- and sequencing platform-specific fusion primers with general, interchangeable barcoding primers, enabling nearly limitless customized barcode-primer combinations. Compared to barcoding with long fusion primers, our multiple-target gene approach is more economical because it overall requires lower number of primers and is based on short primers with generally lower synthesis and purification costs. To highlight our approach, we pooled over 900 different small-subunit rRNA and functional gene amplicon libraries obtained from various environmental or host-associated microbial community samples into a single, paired-end Illumina MiSeq run. Although the amplicon regions ranged in size from approximately 290 to 720 bp, we found no significant systematic sequencing bias related to amplicon length or gene target. Our results indicate that this flexible multiplexing approach produces large, diverse, and high quality sets of amplicon sequence data for modern studies in microbial ecology.
Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells
2015 - Proc Natl Acad Sci USA, 112: E194-203
Abstract:
Microbial communities are essential to the function of virtually all ecosystems and eukaryotes, including humans. However, it is still a major challenge to identify microbial cells active under natural conditions in complex systems. In this study, we developed a new method to identify and sort active microbes on the single-cell level in complex samples using stable isotope probing with heavy water (D2O) combined with Raman microspectroscopy. Incorporation of D2O-derived D into the biomass of autotrophic and heterotrophic bacteria and archaea could be unambiguously detected via C-D signature peaks in single-cell Raman spectra, and the obtained labeling pattern was confirmed by nanoscale-resolution secondary ion MS. In fast-growing Escherichia coli cells, label detection was already possible after 20 min. For functional analyses of microbial communities, the detection of D incorporation from D2O in individual microbial cells via Raman microspectroscopy can be directly combined with FISH for the identification of active microbes. Applying this approach to mouse cecal microbiota revealed that the host-compound foragers Akkermansia muciniphila and Bacteroides acidifaciens exhibited distinctive response patterns to amendments of mucin and sugars. By Raman-based cell sortingof active (deuterated) cells with optical tweezers and subsequent multiple displacement amplification and DNA sequencing, novel cecal microbes stimulated by mucin and/or glucosamine were identified, demonstrating the potential of the nondestructive D2O-Raman approach for targeted sortingof microbial cells with defined functional properties for single-cell genomics.