Publications
Publications in peer reviewed journals
Omics research on abalone (Haliotis spp.): Current state and perspectives
2022 - Aquaculture, 547: 737438
Abstract:
The steady increase in abalone aquaculture production throughout the world has attracted growing interest in the application of new technologies, such as omics approaches for abalone research. Many omics techniques, such as genomics, transcriptomics, proteomics, and metabolomics are becoming established in abalone research and are beginning to reveal key molecules and pathways underlying many biological processes, and to identify associated candidate biomarkers of biological or environmental processes. In this contribution, we synthesize the published omics studies on abalone to highlight the current state of knowledge, open questions, and future directions. In addition, we outline the challenges and limitations of each omics field, some of which could be overcome by integrating multiple omics approaches – a future strategy with great potential for contributing to improve abalone production. Full text
Natural experiments and long-term monitoring are critical to understand and predict marine host-microbe ecology and evolution.
2021 - PLoS Biol, 8: e3001322
Abstract:
Marine multicellular organisms host a diverse collection of bacteria, archaea, microbial eukaryotes, and viruses that form their microbiome. Such host-associated microbes can significantly influence the host's physiological capacities; however, the identity and functional role(s) of key members of the microbiome ("core microbiome") in most marine hosts coexisting in natural settings remain obscure. Also unclear is how dynamic interactions between hosts and the immense standing pool of microbial genetic variation will affect marine ecosystems' capacity to adjust to environmental changes. Here, we argue that significantly advancing our understanding of how host-associated microbes shape marine hosts' plastic and adaptive responses to environmental change requires (i) recognizing that individual host-microbe systems do not exist in an ecological or evolutionary vacuum and (ii) expanding the field toward long-term, multidisciplinary research on entire communities of hosts and microbes. Natural experiments, such as time-calibrated geological events associated with well-characterized environmental gradients, provide unique ecological and evolutionary contexts to address this challenge. We focus here particularly on mutualistic interactions between hosts and microbes, but note that many of the same lessons and approaches would apply to other types of interactions.
Global biogeography of chemosynthetic symbionts reveals both localized and globally distributed symbiont groups.
2021 - Proc Natl Acad Sci U S A, 29: in press
Abstract:
In the ocean, most hosts acquire their symbionts from the environment. Due to the immense spatial scales involved, our understanding of the biogeography of hosts and symbionts in marine systems is patchy, although this knowledge is essential for understanding fundamental aspects of symbiosis such as host-symbiont specificity and evolution. Lucinidae is the most species-rich and widely distributed family of marine bivalves hosting autotrophic bacterial endosymbionts. Previous molecular surveys identified location-specific symbiont types that "promiscuously" form associations with multiple divergent cooccurring host species. This flexibility of host-microbe pairings is thought to underpin their global success, as it allows hosts to form associations with locally adapted symbionts. We used metagenomics to investigate the biodiversity, functional variability, and genetic exchange among the endosymbionts of 12 lucinid host species from across the globe. We report a cosmopolitan symbiont species, Thiodiazotropha taylori, associated with multiple lucinid host species. T. taylori has achieved more success at dispersal and establishing symbioses with lucinids than any other symbiont described thus far. This discovery challenges our understanding of symbiont dispersal and location-specific colonization and suggests both symbiont and host flexibility underpin the ecological and evolutionary success of the lucinid symbiosis.
A novel SAR324 bacterium associated with abalone, Haliotis diversicolor
2021 - Aquaculture Research, 52: 1945-1953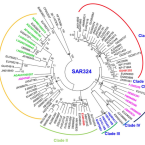
Abstract:
Bacteria affiliated to the Deltaproteobacteria dominate intestinal microbial communities in the abalone, Haliotis diversicolor, and may colonize the host's digestive gland, raising the possibility that they contribute to digestion of macroalgae. However, the phylogenetic, and genomic and metabolic characteristics of these Deltaproteobacteria, and the nature of their relationship to abalone host remain elusive. Here, we examined the intestinal microbial composition of H. diversicolor using high-throughput sequencing and described the genomic characteristics of the Deltaproteobacteria phylotype using genome-centric metagenomics. High-throughput sequencing confirmed that one Deltaproteobacteria phylotype was predominant in intestinal microbiota of H. diversicolor. Phylogeny analysis based on full-length 16S rRNA gene allocated this bacterium to a distinct and unique cluster within SAR324. It possessed a genome of at least 1.59 Mb with 35.15 mol% GC content, much smaller than other sequenced free-living SAR324 bacteria. According to genome annotation and metabolic reconstruction, ATP binding cassette transporters for sugars and carbohydrate metabolism pathways indicated heterotrophic potential. Interestingly, it encoded polysaccharide lyases, which, if expressed, could help the host to digest its macroalgae diet. In contrast to the known SAR324, no sulphur oxidation or carbon fixation pathways were predicted. We propose that this is a unique and specific SAR324 bacterium in symbiosis with Haliotis.
The symbiotic 'all-rounders': Partnerships between marine animals and chemosynthetic nitrogen-fixing bacteria.
2020 - Appl Environ Microbiol, in press
Abstract:
Nitrogen fixation is a widespread metabolic trait in certain types of microorganisms called diazotrophs. Bioavailable nitrogen is limited in various habitats on land and in the sea, and accordingly, a range of plant, animal, and single-celled eukaryotes have evolved symbioses with diverse diazotrophic bacteria, with enormous economic and ecological benefits. Until recently, all known nitrogen-fixing symbionts were heterotrophs such as nodulating rhizobia, or photoautotrophs such as cyanobacteria. In 2016, the first chemoautotrophic nitrogen-fixing symbionts were discovered in a common family of marine clams, the Lucinidae. Chemosynthetic nitrogen-fixing symbionts use the chemical energy stored in reduced sulfur compounds to power carbon and nitrogen fixation, making them metabolic 'all-rounders' with multiple functions in the symbiosis. This distinguishes them from heterotrophic symbionts that require a source of carbon from their host, and their chemosynthetic metabolism distinguishes them from photoautotrophic symbionts that produce oxygen, a potent inhibitor of nitrogenase. In this review, we consider evolutionary aspects of this discovery, by comparing strategies that have evolved for hosting intracellular nitrogen-fixing symbionts in plants and animals. The symbiosis between lucinid clams and chemosynthetic nitrogen-fixing bacteria also has important ecological impacts, as they form a nested symbiosis with endangered marine seagrasses. Notably, nitrogen fixation by lucinid symbionts may help support seagrass health by providing a source of nitrogen in seagrass habitats. These discoveries were enabled by new techniques for understanding the activity of microbial populations in natural environments. However, an animal (or plant) host represents a diverse landscape of microbial niches due to its structural, chemical, immune and behavioural properties. In future, methods that resolve microbial activity at the single cell level will provide radical new insights into the regulation of nitrogen fixation in chemosynthetic symbionts, shedding new light on the evolution of nitrogen-fixing symbioses in contrasting hosts and environments.
A novel alphaproteobacterium with a small genome identified from the digestive gland of multiple species of abalone.
2020 - Environ Microbiol Rep, 4: 387-395
Abstract:
We identified an alphaproteobacterium in the digestive gland of the abalone species Haliotis discus hannai. This phylotype dominated our 16S rRNA clone libraries from the digestive gland of H. discus hannai. Diversity surveys revealed that this phylotype was associated with H. discus hannai and also in another host species, H. gigantea. Whole genome phylogenies placed this bacterium as a new member affiliated with the family Rhodospirillaceae in Alphaproteobacteria. Gene annotation revealed a nearly complete glycolysis pathway but no TCA cycle, but the presence of anaerobic ribonucleoside-triphosphate reductase and oxygen-insensitive NAD(P)H-dependent nitroreductase, which show the genomic potential for anaerobic metabolism. A large cluster of genes encoding ankyrin repeat proteins (ANK) of eukaryotic-like repeat domains and a large gene set for the flagellar system were also detected. Alginate-binding periplasmic proteins and key genes responsible for alginate assimilation were found in the genome, which could potentially contribute to the breakdown of the host's alginate-rich macroalgal diet. These results raise the possibility that this novel alphaproteobacterium is a widespread member of the abalone microbiome that may use polysaccharides derived from its host's macroalgal diet.
Horizontal acquisition of a patchwork Calvin cycle by symbiotic and free-living Campylobacterota (formerly Epsilonproteobacteria).
2020 - ISME J, 1: 104-122
Abstract:
Most autotrophs use the Calvin-Benson-Bassham (CBB) cycle for carbon fixation. In contrast, all currently described autotrophs from the Campylobacterota (previously Epsilonproteobacteria) use the reductive tricarboxylic acid cycle (rTCA) instead. We discovered campylobacterotal epibionts ("Candidatus Thiobarba") of deep-sea mussels that have acquired a complete CBB cycle and may have lost most key genes of the rTCA cycle. Intriguingly, the phylogenies of campylobacterotal CBB cycle genes suggest they were acquired in multiple transfers from Gammaproteobacteria closely related to sulfur-oxidizing endosymbionts associated with the mussels, as well as from Betaproteobacteria. We hypothesize that "Ca. Thiobarba" switched from the rTCA cycle to a fully functional CBB cycle during its evolution, by acquiring genes from multiple sources, including co-occurring symbionts. We also found key CBB cycle genes in free-living Campylobacterota, suggesting that the CBB cycle may be more widespread in this phylum than previously known. Metatranscriptomics and metaproteomics confirmed high expression of CBB cycle genes in mussel-associated "Ca. Thiobarba". Direct stable isotope fingerprinting showed that "Ca. Thiobarba" has typical CBB signatures, suggesting that it uses this cycle for carbon fixation. Our discovery calls into question current assumptions about the distribution of carbon fixation pathways in microbial lineages, and the interpretation of stable isotope measurements in the environment.
Organ transcriptomes of the lucinid clam Loripes orbiculatus (Poli, 1791) provide insights into their specialised roles in the biology of a chemosymbiotic bivalve.
2019 - BMC Genomics, 1: 820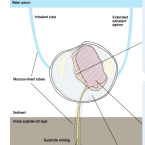
Abstract:
The lucinid clam Loripes orbiculatus lives in a nutritional symbiosis with sulphur-oxidizing bacteria housed in its gills. Although our understanding of the lucinid endosymbiont physiology and metabolism has made significant progress, relatively little is known about how the host regulates the symbiosis at the genetic and molecular levels. We generated transcriptomes from four L. orbiculatus organs (gills, foot, visceral mass, and mantle) for differential expression analyses, to better understand this clam's physiological adaptations to a chemosymbiotic lifestyle, and how it regulates nutritional and immune interactions with its symbionts.
The transcriptome profile of the symbiont-housing gill suggests the regulation of apoptosis and innate immunity are important processes in this organ. We also identified many transcripts encoding ion transporters from the solute carrier family that possibly allow metabolite exchange between host and symbiont. Despite the clam holobiont's clear reliance on chemosynthesis, the clam's visceral mass, which contains the digestive tract, is characterised by enzymes involved in digestion, carbohydrate recognition and metabolism, suggesting that L. orbiculatus has a mixotrophic diet. The foot transcriptome is dominated by the biosynthesis of glycoproteins for the construction of mucus tubes, and receptors that mediate the detection of chemical cues in the environment.
The transcriptome profiles of gills, mantle, foot and visceral mass provide insights into the molecular basis underlying the functional specialisation of bivalve organs adapted to a chemosymbiotic lifestyle.Functional diversity enables multiple symbiont strains to coexist in deep-sea mussels.
2019 - Nat Microbiol, 12: 2487-2497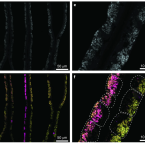
Abstract:
Genetic diversity of closely related free-living microorganisms is widespread and underpins ecosystem functioning, but most evolutionary theories predict that it destabilizes intimate mutualisms. Accordingly, strain diversity is assumed to be highly restricted in intracellular bacteria associated with animals. Here, we sequenced metagenomes and metatranscriptomes of 18 Bathymodiolus mussel individuals from four species, covering their known distribution range at deep-sea hydrothermal vents in the Atlantic. We show that as many as 16 strains of intracellular, sulfur-oxidizing symbionts coexist in individual Bathymodiolus mussels. Co-occurring symbiont strains differed extensively in key functions, such as the use of energy and nutrient sources, electron acceptors and viral defence mechanisms. Most strain-specific genes were expressed, highlighting their potential to affect fitness. We show that fine-scale diversity is pervasive in Bathymodiolus sulfur-oxidizing symbionts, and hypothesize that it may be widespread in low-cost symbioses where the environment, rather than the host, feeds the symbionts.
Horizontally transmitted symbiont populations in deep-sea mussels are genetically isolated.
2019 - ISME J, 12: 2954-2968
Abstract:
Eukaryotes are habitats for bacterial organisms where the host colonization and dispersal among individual hosts have consequences for the bacterial ecology and evolution. Vertical symbiont transmission leads to geographic isolation of the microbial population and consequently to genetic isolation of microbiotas from individual hosts. In contrast, the extent of geographic and genetic isolation of horizontally transmitted microbiota is poorly characterized. Here we show that chemosynthetic symbionts of individual Bathymodiolus brooksi mussels constitute genetically isolated subpopulations. The reconstruction of core genome-wide strains from high-resolution metagenomes revealed distinct phylogenetic clades. Nucleotide diversity and strain composition vary along the mussel life span and individual hosts show a high degree of genetic isolation. Our results suggest that the uptake of environmental bacteria is a restricted process in B. brooksi, where self-infection of the gill tissue results in serial founder effects during symbiont evolution. We conclude that bacterial colonization dynamics over the host life cycle is thus an important determinant of population structure and genome evolution of horizontally transmitted symbionts.
Chemosymbiotic bivalves contribute to the nitrogen budget of seagrass ecosystems.
2019 - ISME J, 12: 3131-3134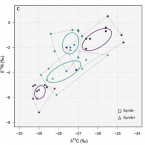
Abstract:
In many seagrass sediments, lucinid bivalves and their sulfur-oxidizing symbionts are thought to underpin key ecosystem functions, but little is known about their role in nutrient cycles, particularly nitrogen. We used natural stable isotopes, elemental analyses, and stable isotope probing to study the ecological stoichiometry of a lucinid symbiosis in spring and fall. Chemoautotrophy appeared to dominate in fall, when chemoautotrophic carbon fixation rates were up to one order of magnitude higher as compared with the spring, suggesting a flexible nutritional mutualism. In fall, an isotope pool dilution experiment revealed carbon limitation of the symbiosis and ammonium excretion rates up to tenfold higher compared with fluxes reported for nonsymbiotic marine bivalves. These results provide evidence that lucinid bivalves can contribute substantial amounts of ammonium to the ecosystem. Given the preference of seagrasses for this nitrogen source, lucinid bivalves' contribution may boost productivity of these important blue carbon ecosystems.
Microbiomes : Importance of Invertebrates in Understanding the Natural Variety of Animal-Microbe Interactions.
2018 - mSystems, 2: in press
Abstract:
Animals evolved in a world teeming with microbes, which play pivotal roles in their health, development, and evolution. Although the overwhelming majority of living animals are invertebrates, the minority of "microbiome" studies focus on this group. Interest in invertebrate-microbe interactions is 2-fold-a range of immune components are conserved across almost all animal (including human) life, and their functional roles may be conserved. Thus, understanding cross talk between microbes and invertebrate animals can lead to insights of broader relevance. Invertebrates offer unique opportunities to "eavesdrop" on intricate host-microbe conversations because they tend to associate with fewer microbes. On the other hand, considering the vast diversity of form and function that has evolved in the invertebrates, they likely evolved an equally diverse range of ways to interact with beneficial microbes. We have investigated only a few of these interactions in detail; thus, there is still great potential for fundamentally new discoveries.
Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis.
2017 - ISME J, 11: 463–477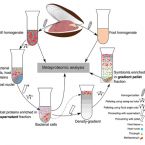
Abstract:
The hydrothermal vent mussel Bathymodiolus azoricus lives in an intimate symbiosis with two types of chemosynthetic Gammaproteobacteria in its gills: a sulfur oxidizer and a methane oxidizer. Despite numerous investigations over the last decades, the degree of interdependence between the three symbiotic partners, their individual metabolic contributions, as well as the mechanism of carbon transfer from the symbionts to the host are poorly understood. We used a combination of proteomics and genomics to investigate the physiology and metabolism of the individual symbiotic partners. Our study revealed that key metabolic functions are most likely accomplished jointly by B. azoricus and its symbionts: (1) CO2 is pre-concentrated by the host for carbon fixation by the sulfur-oxidizing symbiont, and (2) the host replenishes essential biosynthetic TCA cycle intermediates for the sulfur-oxidizing symbiont. In return (3), the sulfur oxidizer may compensate for the host's putative deficiency in amino acid and cofactor biosynthesis. We also identified numerous 'symbiosis-specific' host proteins by comparing symbiont-containing and symbiont-free host tissues and symbiont fractions. These proteins included a large complement of host digestive enzymes in the gill that are likely involved in symbiont digestion and carbon transfer from the symbionts to the host.The ISME Journal advance online publication, 1 November 2016; doi:10.1038/ismej.2016.124.
Ecology and Fisheries: Dark Carbon on Your Dinner Plate.
2016 - Curr. Biol., 24: R1277-R1279
Abstract:
Chemosynthetic primary production by symbiotic microbes powers entire ecosystems in the remote deep sea. New research shows that in shallow waters chemosynthetic symbioses can contribute substantially to a vital economic resource - lobster fisheries in the Caribbean Sea.
Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation.
2016 - Nat Microbiol, 2: 16195
Abstract:
Chemosynthetic symbioses are partnerships between invertebrate animals and chemosynthetic bacteria. The latter are the primary producers, providing most of the organic carbon needed for the animal host's nutrition. We sequenced genomes of the chemosynthetic symbionts from the lucinid bivalve Loripes lucinalis and the stilbonematid nematode Laxus oneistus. The symbionts of both host species encoded nitrogen fixation genes. This is remarkable as no marine chemosynthetic symbiont was previously known to be capable of nitrogen fixation. We detected nitrogenase expression by the symbionts of lucinid clams at the transcriptomic and proteomic level. Mean stable nitrogen isotope values of Loripes lucinalis were within the range expected for fixed atmospheric nitrogen, further suggesting active nitrogen fixation by the symbionts. The ability to fix nitrogen may be widespread among chemosynthetic symbioses in oligotrophic habitats, where nitrogen availability often limits primary productivity.
A specific and widespread association between deep-sea Bathymodiolus mussels and a novel family of Epsilonproteobacteria.
2016 - Environ Microbiol Rep, 8: 805-813
Abstract:
Bathymodiolus mussels dominate animal communities at many hydrothermal vents and cold seeps. Essential to the mussels' ecological and evolutionary success is their association with symbiotic methane- and sulfur-oxidizing gammaproteobacteria, which provide them with nutrition. In addition to these well-known gammaproteobacterial endosymbionts, we found epsilonproteobacterial sequences in metatranscriptomes, metagenomes and 16S rRNA clone libraries as well as by polymerase chain reaction screening of Bathymodiolus species sampled from vents and seeps around the world. These epsilonproteobacterial sequences were closely related, indicating that the association is highly specific. The Bathymodiolus-associated epsilonproteobacterial 16S rRNA sequences were at most 87.6% identical to the closest cultured relative, and 91.2% identical to the closest sequences in public databases. This clade therefore represents a novel family within the Epsilonproteobacteria. Fluorescence in situ hybridization and transmission electron microscopy showed that the bacteria are filamentous epibionts associated with the gill epithelia in two Bathymodiolus species. In animals that host highly specific symbioses with one or a few types of endosymbionts, other less-abundant members of the microbiota can be easily overlooked. Our work highlights how widespread and specific associations with less-abundant microbes can be. Possibly, these microbes play an important role in the survival and health of their animal hosts.
Biophysical and Population Genetic Models Predict the Presence of “Phantom” Stepping Stones Connecting Mid-Atlantic Ridge Vent Ecosystems
2016 - Current Biology, 26: 1 - 11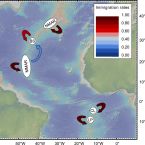
Abstract:
Deep-sea hydrothermal vents are patchily distributed ecosystems inhabited by specialized animal populations that are textbook meta-populations. Many vent-associated species have free-swimming, dispersive larvae that can establish connections between remote populations. However, connectivity patterns among hydrothermal vents are still poorly understood because the deep sea is undersampled, the molecular tools used to date are of limited resolution, and larval dispersal is difficult to measure directly. A better knowledge of connectivity is urgently needed to develop sound environmental management plans for deep-sea mining. Here, we investigated larval dispersal and contemporary connectivity of ecologically important vent mussels (Bathymodiolus spp.) from the Mid-Atlantic Ridge by using high-resolution ocean modeling and population genetic methods. Even when assuming a long pelagic larval duration, our physical model of larval drift suggested that arrival at localities more than 150 km from the source site is unlikely and that dispersal between populations requires intermediate habitats (“phantom” stepping stones). Dispersal patterns showed strong spatiotemporal variability, making predictions of population connectivity challenging. The assumption that mussel populations are only connected via additional stepping stones was supported by contemporary migration rates based on neutral genetic markers. Analyses of population structure confirmed the presence of two southern and two hybridizing northern mussel lineages that exhibited a substantial, though incomplete, genetic differentiation. Our study provides insights into how vent animals can disperse between widely separated vent habitats and shows that recolonization of perturbed vent sites will be subject to chance events, unless connectivity is explicitly considered in the selection of conservation areas.
Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels.
2015 - Elife, e07966
Abstract:
Bathymodiolus mussels live in symbiosis with intracellular sulfur-oxidizing (SOX) bacteria that provide them with nutrition. We sequenced the SOX symbiont genomes from two Bathymodiolus species. Comparison of these symbiont genomes with those of their closest relatives revealed that the symbionts have undergone genome rearrangements, and up to 35% of their genes may have been acquired by horizontal gene transfer. Many of the genes specific to the symbionts were homologs of virulence genes. We discovered an abundant and diverse array of genes similar to insecticidal toxins of nematode and aphid symbionts, and toxins of pathogens such as Yersinia and Vibrio. Transcriptomics and proteomics revealed that the SOX symbionts express the toxin-related genes (TRGs) in their hosts. We hypothesize that the symbionts use these TRGs in beneficial interactions with their host, including protection against parasites. This would explain why a mutualistic symbiont would contain such a remarkable 'arsenal' of TRGs.
Dual symbiosis with co-occurring sulfur-oxidizing symbionts in vestimentiferan tubeworms from a Mediterranean hydrothermal vent.
2014 - Environ. Microbiol., 12: 3638-56
Abstract:
Vestimentiferan Tws colonize hydrothermal vents and cold seeps worldwide. They lack a digestive system and gain nutrition from endosymbiotic sulfur-oxidizing bacteria. It is currently assumed that vestimentiferan Tws harbour only a single endosymbiont type. A few studies found indications for additional symbionts, but conclusive evidence for a multiple symbiosis is still missing. We investigated Tws from Marsili Seamount, a hydrothermal vent in the Mediterranean Sea. Molecular and morphological analyses identified the Tws as Lamellibrachia anaximandri. 16S ribosomal RNA clone libraries revealed two distinct gammaproteobacterial phylotypes that were closely related to sequences from other Lamellibrachia symbionts. Catalysed reporter deposition fluorescence in situ hybridization with specific probes showed that these sequences are from two distinct symbionts. We also found two variants of key genes for sulfur oxidation and carbon fixation, suggesting that both symbiont types are autotrophic sulfur oxidizers. Our results therefore show that vestimentiferans can host multiple co-occurring symbiont types. Statistical analyses of vestimentiferan symbiont diversity revealed that host genus, habitat type, water depth and geographic region together accounted for 27% of genetic diversity, but only water depth had a significant effect on its own. Phylogenetic analyses showed a clear grouping of sequences according to depth, thus confirming the important role water depth played in shaping vestimentiferan symbiont diversity.
The gill chamber epibiosis of deep-sea shrimp Rimicaris exoculata: an in-depth metagenomic investigation and discovery of Zetaproteobacteria.
2014 - Environ. Microbiol., 9: 2723-38
Abstract:
The gill chamber of deep-sea hydrothermal vent shrimp Rimicaris exoculata hosts a dense community of epibiotic bacteria dominated by filamentous Epsilonproteobacteria and Gammaproteobacteria. Using metagenomics on shrimp from the Rainbow hydrothermal vent field, we showed that both epibiont groups have the potential to grow autotrophically and oxidize reduced sulfur compounds or hydrogen with oxygen or nitrate. For carbon fixation, the Epsilonproteobacteria use the reductive tricarboxylic acid cycle, whereas the Gammaproteobacteria use the Calvin-Benson-Bassham cycle. Only the epsilonproteobacterial epibionts had the genes necessary for producing ammonium. This ability likely minimizes direct competition between epibionts and also broadens the spectrum of environmental conditions that the shrimp may successfully inhabit. We identified genes likely to be involved in shrimp-epibiont interactions, as well as genes for nutritional and detoxification processes that might benefit the host. Shrimp epibionts at Rainbow are often coated with iron oxyhydroxides, whose origin is intensely debated. We identified 16S rRNA sequences and functional genes affiliated with iron-oxidizing Zetaproteobacteria, which indicates that biological iron oxidation might play a role in forming these deposits. Fluorescence in situ hybridizations confirmed the presence of active Zetaproteobacteria in the R. exoculata gill chamber, thus providing the first evidence for a Zetaproteobacteria-invertebrate association.
Bacterial symbionts of Bathymodiolus mussels and Escarpia tubeworms from Chapopote, an asphalt seep in the Southern Gulf of Mexico.
2013 - Environ. Microbiol., 7: 1969-87
Abstract:
Chemosynthetic life was recently discovered at Chapopote, an asphalt hydrocarbon seep in the southern Gulf of Mexico. Preliminary morphological analyses indicated that one tubeworm and two mussel species colonize Chapopote. Our molecular analyses identified the tubeworm as Escarpia sp., and the mussels as Bathymodiolus heckerae and B. brooksi. Comparative 16S rRNA analysis and FISH showed that all three species harbour intracellular sulfur-oxidizing symbionts highly similar or identical to those found in the same host species from northern Gulf of Mexico (nGoM). The mussels also harbour methane-oxidizing symbionts, and these shared highly similar to identical 16S rRNA sequences to their nGoM conspecifics. We discovered a novel symbiont in B. heckerae, which is closely related to hydrocarbon-degrading bacteria of the genus Cycloclasticus. In B. heckerae, we found key genes for the use of aromatic compounds, and its stable carbon isotope values were consistently higher than B. brooksi, indicating that the novel symbiont might use isotopically heavy aromatic hydrocarbons from the asphalt seep. This discovery is particularly intriguing because until now only methane and reduced sulfur compounds have been shown to power cold-seep chemosynthetic symbioses. The abundant hydrocarbons available at Chapopote would provide these mussel symbioses with a rich source of nutrition.
Origins and evolutionary flexibility of chemosynthetic symbionts from deep-sea animals.
2012 - Biol. Bull., 1: 123-37
Abstract:
Bathymodiolin mussels dominate hydrothermal vent and cold seep communities worldwide. Symbiotic associations with chemosynthetic sulfur- and methane-oxidizing bacteria that provide for their nutrition are the key to their ecological and evolutionary success. The current paradigm is that these symbioses evolved from two free-living ancestors, one methane-oxidizing and one sulfur-oxidizing bacterium. In contrast to previous studies, our phylogenetic analyses of the bathymodiolin symbionts show that both the sulfur and the methane oxidizers fall into multiple clades interspersed with free-living bacteria, many of which were discovered recently in metagenomes from marine oxygen minimum zones. We therefore hypothesize that symbioses between bathymodiolin mussels and free-living sulfur- and methane-oxidizing bacteria evolved multiple times in convergent evolution. Furthermore, by 16S rRNA sequencing and fluorescence in situ hybridization, we show that close relatives of the bathymodiolin symbionts occur on hosts belonging to different animal phyla: Raricirrus beryli, a terebellid polychaete from a whale-fall, and a poecilosclerid sponge from a cold seep. The host range within the bathymodiolin symbionts is therefore greater than previously recognized, confirming the remarkable flexibility of these symbiotic associations.
Convergent and divergent evolution of metabolism in sulfur-oxidizing symbionts and the role of horizontal gene transfer.
2012 - Curr. Opin. Microbiol., 5: 621-31
Abstract:
Symbioses between marine invertebrates and autotrophic sulfur-oxidizing bacteria have evolved from multiple lineages within the Gammaproteobacteria in a striking example of convergent evolution. These GammaSOX symbionts all perform the same basic function: they provide their hosts with nutrition through the fixation of CO(2) into biomass using reduced sulfur compounds as an energy source. However, our review of recent -omics based studies and genome mining for this study revealed that the GammaSOX symbionts diverge in many other metabolic capabilities and functions, and we show how these divergences could reflect adaptations to different hosts and habitat conditions. Our phylogenetic analyses of key metabolic genes in GammaSOX symbionts revealed that these differed markedly from 16S rRNA phylogenies. We hypothesize that horizontal gene transfer (HGT) would explain many of these incongruencies, and conclude that HGT may have played a significant role in shaping the metabolic evolution of GammaSOX symbionts.
Genetic connectivity between north and south Mid-Atlantic Ridge chemosynthetic bivalves and their symbionts.
2012 - PLoS One, 7: e39994
Abstract:
Transform faults are geological structures that interrupt the continuity of mid-ocean ridges and can act as dispersal barriers for hydrothermal vent organisms. In the equatorial Atlantic Ocean, it has been hypothesized that long transform faults impede gene flow between the northern and the southern Mid-Atlantic Ridge (MAR) and disconnect a northern from a southern biogeographic province. To test if there is a barrier effect in the equatorial Atlantic, we examined phylogenetic relationships of chemosynthetic bivalves and their bacterial symbionts from the recently discovered southern MAR hydrothermal vents at 5°S and 9°S. We examined Bathymodiolus spp. mussels and Abyssogena southwardae clams using the mitochondrial cytochrome c oxidase subunit I (COI) gene as a phylogenetic marker for the hosts and the bacterial 16S rRNA gene as a marker for the symbionts. Bathymodiolus spp. from the two southern sites were genetically divergent from the northern MAR species B. azoricus and B. puteoserpentis but all four host lineages form a monophyletic group indicating that they radiated after divergence from their northern Atlantic sister group, the B. boomerang species complex. This suggests dispersal of Bathymodiolus species from north to south across the equatorial belt. 16S rRNA genealogies of chemoautotrophic and methanotrophic symbionts of Bathymodiolus spp. were inconsistent and did not match the host COI genealogy indicating disconnected biogeography patterns. The vesicomyid clam Abyssogena southwardae from 5°S shared an identical COI haplotype with A. southwardae from the Logatchev vent field on the northern MAR and their symbionts shared identical 16S phylotypes, suggesting gene flow across the Equator. Our results indicate genetic connectivity between the northern and southern MAR and suggest that a strict dispersal barrier does not exist.
Pathways of carbon and energy metabolism of the epibiotic community associated with the deep-sea hydrothermal vent shrimp Rimicaris exoculata.
2011 - PLoS One, 1: e16018
Abstract:
The shrimp Rimicaris exoculata dominates the faunal biomass at many deep-sea hydrothermal vent sites at the Mid-Atlantic Ridge. In its enlarged gill chamber it harbors a specialized epibiotic bacterial community for which a nutritional role has been proposed.
We analyzed specimens from the Snake Pit hydrothermal vent field on the Mid-Atlantic Ridge by complementing a 16S rRNA gene survey with the analysis of genes involved in carbon, sulfur and hydrogen metabolism. In addition to Epsilon- and Gammaproteobacteria, the epibiotic community unexpectedly also consists of Deltaproteobacteria of a single phylotype, closely related to the genus Desulfocapsa. The association of these phylogenetic groups with the shrimp was confirmed by fluorescence in situ hybridization. Based on functional gene analyses, we hypothesize that the Gamma- and Epsilonproteobacteria are capable of autotrophic growth by oxidizing reduced sulfur compounds, and that the Deltaproteobacteria are also involved in sulfur metabolism. In addition, the detection of proteobacterial hydrogenases indicates the potential for hydrogen oxidation in these communities. Interestingly, the frequency of these phylotypes in 16S rRNA gene clone libraries from the mouthparts differ from that of the inner lining of the gill chamber, indicating potential functional compartmentalization.
Our data show the specific association of autotrophic bacteria with Rimicaris exoculata from the Snake Pit hydrothermal vent field, and suggest that autotrophic carbon fixation is contributing to the productivity of the epibiotic community with the reductive tricarboxylic acid cycle as one important carbon fixation pathway. This has not been considered in previous studies of carbon fixation and stable carbon isotope composition of the shrimp and its epibionts. Furthermore, the co-occurrence of sulfur-oxidizing and sulfur-reducing epibionts raises the possibility that both may be involved in the syntrophic exchange of sulfur compounds, which could increase the overall efficiency of this epibiotic community.Hydrogen is an energy source for hydrothermal vent symbioses.
2011 - Nature, 7359: 176-80
Abstract:
The discovery of deep-sea hydrothermal vents in 1977 revolutionized our understanding of the energy sources that fuel primary productivity on Earth. Hydrothermal vent ecosystems are dominated by animals that live in symbiosis with chemosynthetic bacteria. So far, only two energy sources have been shown to power chemosynthetic symbioses: reduced sulphur compounds and methane. Using metagenome sequencing, single-gene fluorescence in situ hybridization, immunohistochemistry, shipboard incubations and in situ mass spectrometry, we show here that the symbionts of the hydrothermal vent mussel Bathymodiolus from the Mid-Atlantic Ridge use hydrogen to power primary production. In addition, we show that the symbionts of Bathymodiolus mussels from Pacific vents have hupL, the key gene for hydrogen oxidation. Furthermore, the symbionts of other vent animals such as the tubeworm Riftia pachyptila and the shrimp Rimicaris exoculata also have hupL. We propose that the ability to use hydrogen as an energy source is widespread in hydrothermal vent symbioses, particularly at sites where hydrogen is abundant.
Geochemical constraints on the diversity and activity of H2 -oxidizing microorganisms in diffuse hydrothermal fluids from a basalt- and an ultramafic-hosted vent.
2010 - FEMS Microbiol. Ecol., 1: 55-71
Abstract:
Mixing processes of reduced hydrothermal fluids with oxygenated seawater and fluid-rock reactions contribute to the chemical signatures of diffuse venting and likely determine the geochemical constraints on microbial life. We examined the influence of fluid chemistry on microbial diversity and activity by sampling diffuse fluids emanating through mussel beds at two contrasting hydrothermal vents. The H(2) concentration was very low at the basalt-hosted Clueless site, and mixing models suggest O(2) availability throughout much of the habitat. In contrast, effluents from the ultramafic-hosted Quest site were considerably enriched in H(2) , while O(2) is likely limited to the mussel layer. Only two different hydrogenase genes were identified in clone libraries from the H(2) -poor Clueless fluids, but these fluids exhibited the highest H(2) uptake rates in H(2) -spiked incubations (oxic conditions, at 18 °C). In contrast, a phylogenetically diverse H(2) -oxidizing potential was associated with distinct thermal conditions in the H(2) -rich Quest fluids, but under oxic conditions, H(2) uptake rates were extremely low. Significant stimulation of CO(2) fixation rates by H(2) addition was solely illustrated in Quest incubations (P-value <0.02), but only in conjunction with anoxic conditions (at 18 °C). We conclude that the factors contributing toward differences in the diversity and activity of H(2) oxidizers at these sites include H(2) and O(2) availability.
Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields.
2010 - Environ. Microbiol., 8: 2204-18
Abstract:
The shrimp Rimicaris exoculata from hydrothermal vents on the Mid-Atlantic Ridge (MAR) harbours bacterial epibionts on specialized appendages and the inner surfaces of its gill chamber. Using comparative 16S rRNA sequence analysis and fluorescence in situ hybridization (FISH), we examined the R. exoculata epibiosis from four vents sites along the known distribution range of the shrimp on the MAR. Our results show that R. exoculata lives in symbiosis with two types of filamentous epibionts. One belongs to the Epsilonproteobacteria, and was previously identified as the dominant symbiont of R. exoculata. The second is a novel gammaproteobacterial symbiont that belongs to a clade consisting exclusively of sequences from epibiotic bacteria of hydrothermal vent animals, with the filamentous sulfur oxidizer Leucothrix mucor as the closest free-living relative. Both the epsilon- and the gammaproteobacterial symbionts dominated the R. exoculata epibiosis at all four MAR vent sites despite striking differences between vent fluid chemistry and distances between sites of up to 8500 km, indicating that the symbiosis is highly stable and specific. Phylogenetic analyses of two mitochondrial host genes showed little to no differences between hosts from the four vent sites. In contrast, there was significant spatial structuring of both the gamma- and the epsilonproteobacterial symbiont populations based on their 16S rRNA gene sequences that was correlated with geographic distance along the MAR. We hypothesize that biogeography and host-symbiont selectivity play a role in structuring the epibiosis of R. exoculata.
Methanotrophic symbioses in marine invertebrates.
2009 - Environ Microbiol Rep, 5: 319-35
Abstract:
Symbioses between marine animals and aerobic methane-oxidizing bacteria are found at hydrothermal vents and cold seeps in the deep sea where reduced, methane-rich fluids mix with the surrounding oxidized seawater. These habitats are 'oases' in the otherwise nutrient-poor deep sea, where entire ecosystems are fueled by microbial chemosynthesis. By associating with bacteria that gain energy from the oxidation of CH4 with O2 , the animal host is indirectly able to gain nutrition from methane, an energy source that is otherwise only available to methanotrophic microorganisms. The host, in turn, provides its symbionts with continuous access to both electron acceptors and donors that are only available at a narrow oxic - anoxic interface for free-living methanotrophs. Symbiotic methane oxidizers have resisted all attempts at cultivation, so that all evidence for these symbiotic associations comes from ultrastructural, enzymatic, physiological, stable isotope and molecular biological studies of the symbiotic host tissues. In this review, we present an overview of the habitats and invertebrate hosts in which symbiotic methane oxidizers have been found, and the methods used to investigate these symbioses, focusing on the symbioses of bathymodiolin mussels that have received the most attention among methanotrophic associations.